Bidirectional linkage of DNA barcodes for the multiplexed mapping of higher-order protein interactions in cells
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
Abstract
Capturing the full complexity of the diverse hierarchical interactions in the protein interactome is challenging. Here we report a DNA-barcoding method for the multiplexed mapping of pairwise and higher-order protein interactions and their dynamics within cells. The method leverages antibodies conjugated with barcoded DNA strands that can bidirectionally hybridize and covalently link to linearize closely spaced interactions within individual 3D protein complexes, encoding and decoding the protein constituents and the interactions among them. By mapping protein interactions in cancer cells and normal cells, we found that tumour cells exhibit a larger diversity and abundance of protein complexes with higher-order interactions. In biopsies of human breast-cancer tissue, the method accurately identified the cancer subtype and revealed that higher-order protein interactions are associated with cancer aggressiveness. A method leveraging antibodies conjugated with barcoded DNA strands to linearize closely spaced interactions from individual 3D protein complexes allows for the mapping of pairwise and higher-order protein interactions within cells.
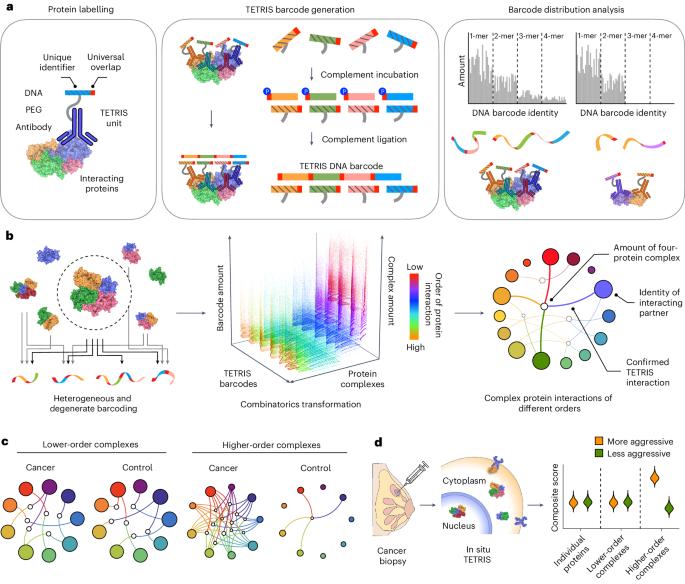
双向连接 DNA 条形码,用于绘制细胞中高阶蛋白质相互作用的复用图谱
捕捉蛋白质相互作用组中不同层次相互作用的全部复杂性具有挑战性。在这里,我们报告了一种 DNA 条形码方法,用于多路复用绘制成对和高阶蛋白质相互作用及其在细胞内的动态。该方法利用抗体与条形码 DNA 链连接,条形码 DNA 链可以双向杂交和共价连接,线性化单个三维蛋白质复合物内紧密间隔的相互作用,编码和解码蛋白质成分及其之间的相互作用。通过绘制癌细胞和正常细胞中蛋白质相互作用的图谱,我们发现肿瘤细胞中具有高阶相互作用的蛋白质复合物的多样性和丰度更高。在人类乳腺癌组织活检中,该方法准确地识别了癌症亚型,并揭示了高阶蛋白相互作用与癌症的侵袭性有关。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


