The HADDOCK2.4 web server for integrative modeling of biomolecular complexes
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Interactions between macromolecules, such as proteins and nucleic acids, are essential for cellular functions. Experimental methods can fail to provide all the information required to fully model biomolecular complexes at atomic resolution, particularly for large and heterogeneous assemblies. Integrative computational approaches have, therefore, gained popularity, complementing traditional experimental methods in structural biology. Here, we introduce HADDOCK2.4, an integrative modeling platform, and its updated web interface ( https://wenmr.science.uu.nl/haddock2.4 ). The platform seamlessly integrates diverse experimental and theoretical data to generate high-quality models of macromolecular complexes. The user-friendly web server offers automated parameter settings, access to distributed computing resources, and pre- and post-processing steps that enhance the user experience. To present the web server’s various interfaces and features, we demonstrate two different applications: (i) we predict the structure of an antibody–antigen complex by using NMR data for the antigen and knowledge of the hypervariable loops for the antibody, and (ii) we perform coarse-grained modeling of PRC1 with a nucleosome particle guided by mutagenesis and functional data. The described protocols require some basic familiarity with molecular modeling and the Linux command shell. This new version of our widely used HADDOCK web server allows structural biologists and non-experts to explore intricate macromolecular assemblies encompassing various molecule types. The HADDOCK2.4 web server is a modeling platform that can integrate experimental and theoretical data for guiding 3D prediction of biomolecular complexes.
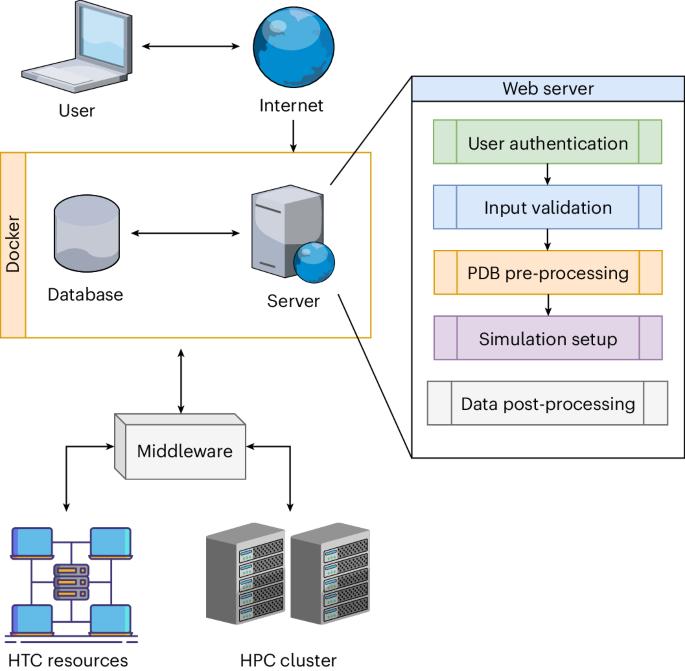
用于生物分子复合物综合建模的 HADDOCK2.4 网络服务器。
蛋白质和核酸等大分子之间的相互作用对细胞功能至关重要。实验方法可能无法提供在原子分辨率下全面模拟生物分子复合物所需的全部信息,尤其是对于大型异质组装体而言。因此,综合计算方法越来越受欢迎,成为结构生物学中传统实验方法的补充。在此,我们介绍 HADDOCK2.4(一种整合建模平台)及其更新的网络界面 ( https://wenmr.science.uu.nl/haddock2.4 )。该平台无缝整合各种实验和理论数据,生成高质量的大分子复合物模型。用户友好的网络服务器提供自动参数设置、访问分布式计算资源以及预处理和后处理步骤,从而增强了用户体验。为了展示网络服务器的各种界面和功能,我们演示了两个不同的应用:(i) 利用抗原的核磁共振数据和抗体超变环的知识预测抗体-抗原复合物的结构;(ii) 在诱变和功能数据的指导下,对带有核小体颗粒的 PRC1 进行粗粒度建模。所描述的协议要求对分子建模和 Linux 命令外壳有一定的基本了解。我们广泛使用的 HADDOCK 网络服务器的新版本允许结构生物学家和非专业人员探索包括各种分子类型的复杂大分子组装。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


