High-throughput single-cell transcriptomics of bacteria using combinatorial barcoding
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
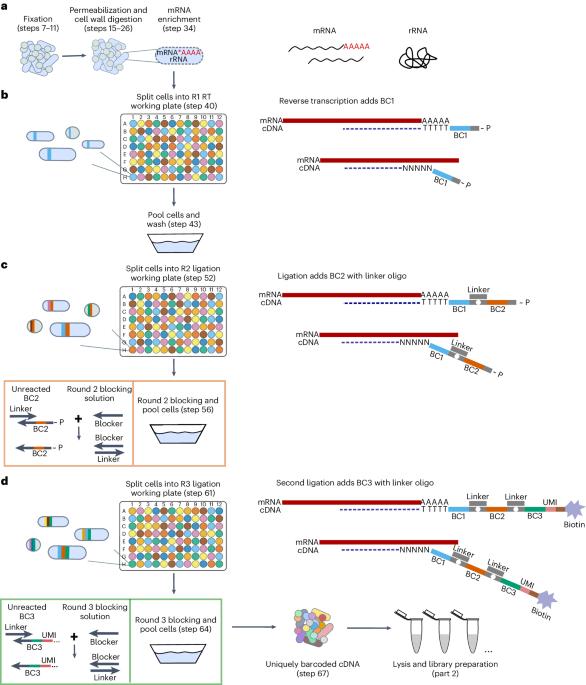
Microbial split-pool ligation transcriptomics (microSPLiT) is a high-throughput single-cell RNA sequencing method for bacteria. With four combinatorial barcoding rounds, microSPLiT can profile transcriptional states in hundreds of thousands of Gram-negative and Gram-positive bacteria in a single experiment without specialized equipment. As bacterial samples are fixed and permeabilized before barcoding, they can be collected and stored ahead of time. During the first barcoding round, the fixed and permeabilized bacteria are distributed into a 96-well plate, where their transcripts are reverse transcribed into cDNA and labeled with the first well-specific barcode inside the cells. The cells are mixed and redistributed two more times into new 96-well plates, where the second and third barcodes are appended to the cDNA via in-cell ligation reactions. Finally, the cells are mixed and divided into aliquot sub-libraries, which can be stored until future use or prepared for sequencing with the addition of a fourth barcode. It takes 4 days to generate sequencing-ready libraries, including 1 day for collection and overnight fixation of samples. The standard plate setup enables single-cell transcriptional profiling of up to 1 million bacterial cells and up to 96 samples in a single barcoding experiment, with the possibility of expansion by adding barcoding rounds. The protocol requires experience in basic molecular biology techniques, handling of bacterial samples and preparation of DNA libraries for next-generation sequencing. It can be performed by experienced undergraduate or graduate students. Data analysis requires access to computing resources, familiarity with Unix command line and basic experience with Python or R. Single-cell transcriptomics of bacteria is challenging. microSPLiT is a high-throughput method for single-cell RNA sequencing of both Gram-positive and Gram-negative bacteria using combinatorial barcoding without the need for specialized equipment.
利用组合条形码对细菌进行高通量单细胞转录组学研究
微生物分离池连接转录组学(microSPLiT)是一种用于细菌的高通量单细胞 RNA 测序方法。通过四轮组合条形码,microSPLiT 可以在一次实验中分析数十万革兰氏阴性和革兰氏阳性细菌的转录状态,而无需专业设备。由于细菌样本在条形码编码前已经固定和渗透,因此可以提前收集和储存。在第一轮条形码编码过程中,固定和渗透的细菌被分装到 96 孔板中,其转录本被反转录成 cDNA,并在细胞内标记上第一井特异性条形码。然后将细胞混合并两次重新分配到新的 96 孔板中,通过细胞内连接反应将第二和第三个条形码添加到 cDNA 中。最后,将细胞混合并分成等分的子库,这些子库可以保存到将来使用,也可以在加入第四个条形码后准备测序。生成可用于测序的文库需要 4 天时间,其中包括 1 天的样本收集和过夜固定时间。标准平板设置可在单次条形码实验中对多达 100 万个细菌细胞和 96 个样本进行单细胞转录分析,并可通过增加条形码轮次进行扩展。该方案需要具备基本的分子生物学技术、处理细菌样本和制备用于下一代测序的 DNA 文库的经验。有经验的本科生或研究生均可完成。数据分析需要使用计算资源、熟悉 Unix 命令行和 Python 或 R 的基本经验。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


