Viral metagenome reveals microbial hosts and the associated antibiotic resistome on microplastics
引用次数: 0
Abstract
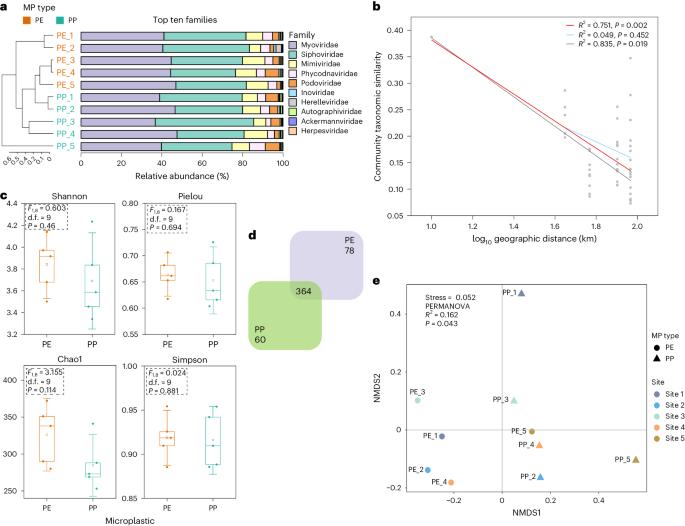
Microplastics provide a unique niche for viruses, promoting viral interactions with hosts and accelerating the rapid ‘horizontal’ spread of antibiotic resistance genes (ARGs). Currently, however, there is a lack of knowledge concerning the main drivers for viral distribution on microplastics and on the resulting patterns of viral biogeographic distributions and the spread of the associated ARGs. Here we performed metagenomic and virus enrichment-based viromic sequencings on both polyethylene and polypropylene microplastics along a river. Experimental results show that Proteobacteria, Firmicutes, Actinobacteria and Cyanobacteria were the potential hosts of viruses on microplastics, but only approximately 4.1% of viral variations were associated with a bacterial community. Notably, two shared ARGs and six metal resistance genes were identified in both viral and their host bacterial genomes, indicating the occurrence of horizontal gene transfer between viruses and bacteria. Furthermore, microplastics introduce more distinctive elements to viral ecology, fostering viral diversification and virus–host linkage while refraining from an escalated level of horizontal gene transfer of ARGs in contrast to natural matrixes. Our study provides comprehensive profiles of viral communities, virus-related ARGs and their driving factors on microplastics, highlighting how these anthropogenic niches provide unique interfaces that comprise highly defined viral ecological features in the environment. The combination of metagenome and virus-enriched virome sequencing provides comprehensive profiles of viral communities and their associated antibiotic resistance genes and reveals the factors driving changes in the plastisphere.
病毒元基因组揭示微塑料上的微生物宿主及相关抗生素耐药性基因组
微塑料为病毒提供了一个独特的生存环境,促进了病毒与宿主的相互作用,并加速了抗生素抗性基因(ARGs)的快速 "水平 "传播。然而,目前人们对微塑料上病毒分布的主要驱动因素、由此产生的病毒生物地理分布模式以及相关 ARGs 的传播还缺乏了解。在此,我们对河流沿岸的聚乙烯和聚丙烯微塑料进行了元基因组学和基于病毒富集的病毒组测序。实验结果表明,变形菌、固着菌、放线菌和蓝细菌是微塑料上病毒的潜在宿主,但只有约 4.1% 的病毒变异与细菌群落有关。值得注意的是,在病毒及其宿主细菌基因组中发现了两个共享的ARGs和六个抗金属基因,这表明病毒和细菌之间存在水平基因转移。此外,微塑料还为病毒生态学引入了更多独特的元素,促进了病毒的多样化和病毒-宿主联系,同时与自然基质相比,ARGs 的水平基因转移没有升级。我们的研究提供了微塑料上病毒群落、与病毒相关的ARGs及其驱动因素的综合概况,强调了这些人为壁龛如何提供独特的界面,构成环境中高度确定的病毒生态特征。元基因组测序与富集病毒的病毒组测序相结合,提供了病毒群落及其相关抗生素耐药基因的全面概况,并揭示了驱动塑界变化的因素。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


