Eosinophil count in cecal biopsies using artificial intelligence-based machine learning
Abstract
Background
Tissue eosinophilia is a diagnostically challenging entity. An accurate diagnosis can benefit the patient with appropriate treatment. Histopathology is the gold standard. Manual counting results in errors and is time consuming. Artificial intelligence (AI) acts like a human brain in terms of memory and reproducibility.
Methods
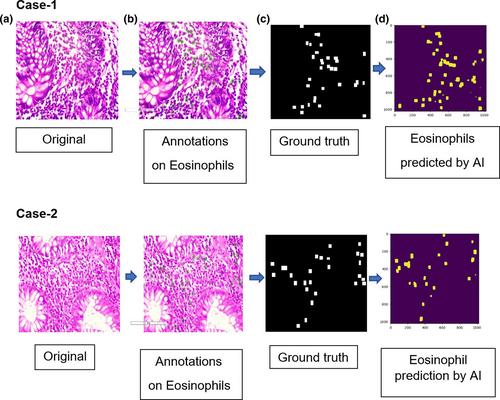
The aim of this study was to determine the eosinophil count in gastrointestinal biopsies using AI and compare it with the manual method. A total of 400 digital images of hematoxylin and eosin-stained slides of gastrointestinal biopsies were included in the study. Annotations were performed on images containing eosinophils. The neural network prepared using the Python language was a modified U-Net architecture containing five down blocks and five up blocks. The results obtained using the AI model were compared with manual counts. Pearson's correlation coefficient and Cohen's kappa coefficient of agreement were used for statistical analysis.
Results
Pearson's correlation coefficient demonstrated a strong positive correlation between AI and manual eosinophil (0.8), which suggests a very strong correlation. The Cohen's kappa coefficient of agreement was 0.85, which corresponds to excellent agreement.
Conclusions
The manual eosinophil count and AI predictions demonstrated a very strong positive correlation. It is necessary to count eosinophils using an AI model because it saves time, and eliminates interobserver variability and human error.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


