Big data and deep learning for RNA biology
IF 12.9
2区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The exponential growth of big data in RNA biology (RB) has led to the development of deep learning (DL) models that have driven crucial discoveries. As constantly evidenced by DL studies in other fields, the successful implementation of DL in RB depends heavily on the effective utilization of large-scale datasets from public databases. In achieving this goal, data encoding methods, learning algorithms, and techniques that align well with biological domain knowledge have played pivotal roles. In this review, we provide guiding principles for applying these DL concepts to various problems in RB by demonstrating successful examples and associated methodologies. We also discuss the remaining challenges in developing DL models for RB and suggest strategies to overcome these challenges. Overall, this review aims to illuminate the compelling potential of DL for RB and ways to apply this powerful technology to investigate the intriguing biology of RNA more effectively. This review spotlights the revolutionary role of deep learning (DL) in expanding the understanding of RNA. RNA is a fundamental biomolecule that shapes and regulates diverse phenotypes including human diseases. Understanding the principles governing the functions of RNA is a key objective of current biology. Recently, big data produced via high-throughput experiments have been utilized to develop DL models aimed at analyzing and predicting RNA-related biological processes. This review emphasizes the role of public databases in providing these big data for training DL models. The authors introduce core DL concepts necessary for training models from the biological data. By extensively examining DL studies in various fields of RNA biology, the authors suggest how to better leverage DL for revealing novel biological knowledge and demonstrate the potential of DL in deciphering the complex biology of RNA. This summary was initially drafted using artificial intelligence, then revised and fact-checked by the author.
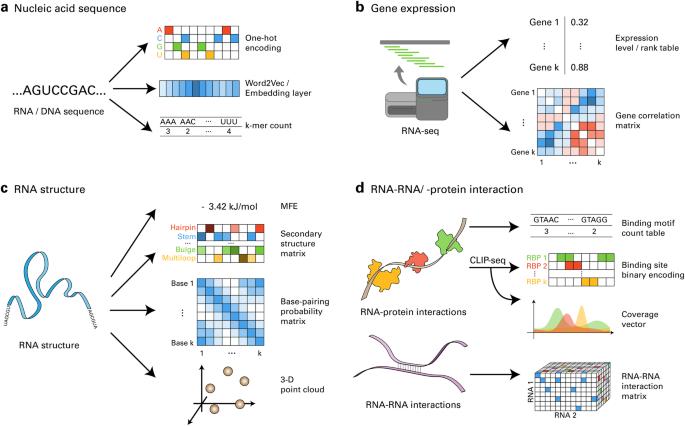
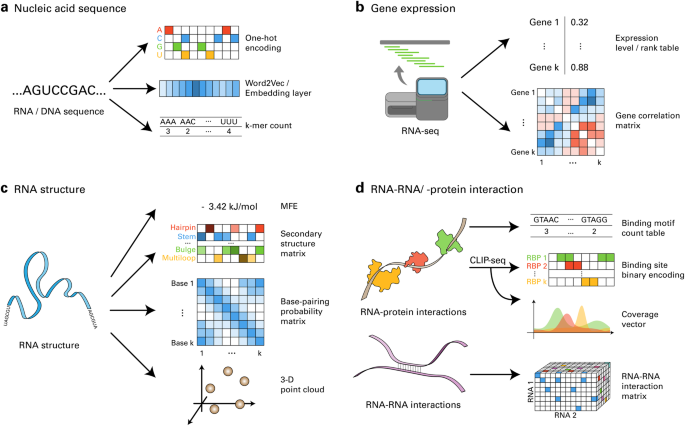
用于 RNA 生物学的大数据和深度学习。
RNA 生物学(RB)领域大数据的指数级增长促进了深度学习(DL)模型的发展,推动了重要发现的产生。正如其他领域的深度学习研究不断证明的那样,在 RB 中成功实施深度学习在很大程度上取决于对来自公共数据库的大规模数据集的有效利用。在实现这一目标的过程中,数据编码方法、学习算法以及与生物领域知识相匹配的技术发挥了关键作用。在本综述中,我们通过展示成功案例和相关方法,为将这些 DL 概念应用于 RB 中的各种问题提供了指导原则。我们还讨论了为遗传资源开发数字语言模型的其余挑战,并提出了克服这些挑战的策略。总之,本综述旨在阐明 DL 在 RB 方面令人瞩目的潜力,以及如何应用这一强大技术更有效地研究 RNA 这一引人入胜的生物学问题。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Experimental and Molecular Medicine
医学-生化与分子生物学
CiteScore
19.50
自引率
0.80%
发文量
166
审稿时长
3 months
期刊介绍:
Experimental & Molecular Medicine (EMM) stands as Korea's pioneering biochemistry journal, established in 1964 and rejuvenated in 1996 as an Open Access, fully peer-reviewed international journal. Dedicated to advancing translational research and showcasing recent breakthroughs in the biomedical realm, EMM invites submissions encompassing genetic, molecular, and cellular studies of human physiology and diseases. Emphasizing the correlation between experimental and translational research and enhanced clinical benefits, the journal actively encourages contributions employing specific molecular tools. Welcoming studies that bridge basic discoveries with clinical relevance, alongside articles demonstrating clear in vivo significance and novelty, Experimental & Molecular Medicine proudly serves as an open-access, online-only repository of cutting-edge medical research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


