Single-cell transcriptomics across 2,534 microbial species reveals functional heterogeneity in the rumen microbiome
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
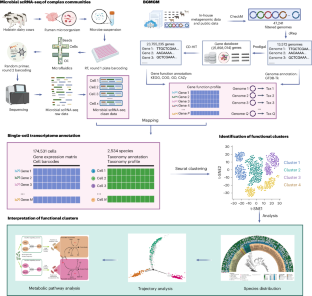
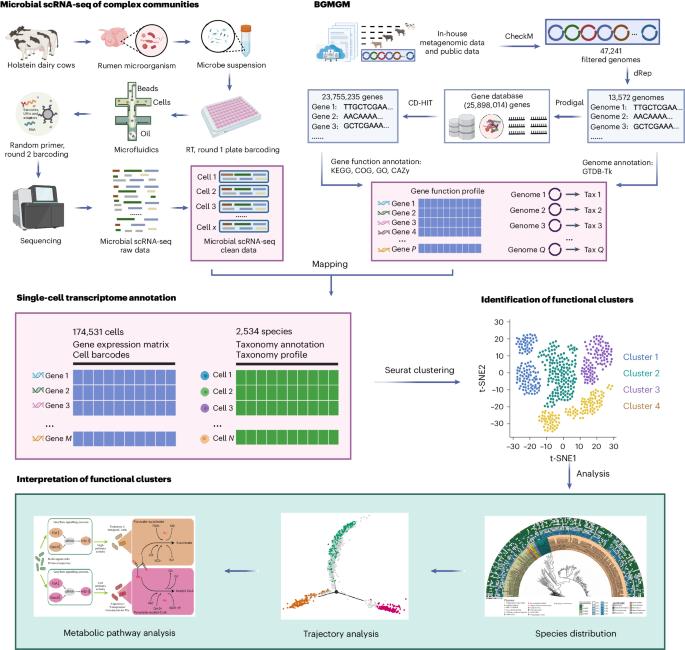
Deciphering the activity of individual microbes within complex communities and environments remains a challenge. Here we describe the development of microbiome single-cell transcriptomics using droplet-based single-cell RNA sequencing and pangenome-based computational analysis to characterize the functional heterogeneity of the rumen microbiome. We generated a microbial genome database (the Bovine Gastro Microbial Genome Map) as a functional reference map for the construction of a single-cell transcriptomic atlas of the rumen microbiome. The atlas includes 174,531 microbial cells and 2,534 species, of which 172 are core active species grouped into 12 functional clusters. We detected single-cell-level functional roles, including a key role for Basfia succiniciproducens in the carbohydrate metabolic niche of the rumen microbiome. Furthermore, we explored functional heterogeneity and reveal metabolic niche trajectories driven by biofilm formation pathway genes within B. succiniciproducens. Our results provide a resource for studying the rumen microbiome and illustrate the diverse functions of individual microbial cells that drive their ecological niche stability or adaptation within the ecosystem. A single-cell transcriptomic resource of 174,531 microbial cells across 2,534 species allows the detection of single-cell-level functional roles in the rumen microbiome.
跨越 2,534 个微生物物种的单细胞转录组学揭示了瘤胃微生物组的功能异质性
破解复杂群落和环境中单个微生物的活动仍然是一项挑战。在此,我们介绍了微生物组单细胞转录组学的发展情况,该方法利用基于液滴的单细胞RNA测序和基于泛基因组的计算分析来描述瘤胃微生物组的功能异质性。我们生成了一个微生物基因组数据库(牛胃肠道微生物基因组图谱),作为构建瘤胃微生物组单细胞转录组图谱的功能参考图谱。该图谱包括 174,531 个微生物细胞和 2,534 个物种,其中 172 个为核心活性物种,分为 12 个功能群组。我们发现了单细胞级的功能作用,其中包括琥珀酰琥珀虫(Basfia succiniciproducens)在瘤胃微生物群碳水化合物代谢生态位中的关键作用。此外,我们还探索了功能异质性,并揭示了琥珀酰琥珀虫体内由生物膜形成途径基因驱动的代谢生态位轨迹。我们的研究结果为研究瘤胃微生物组提供了资源,并说明了单个微生物细胞的不同功能,这些功能驱动着它们在生态系统中的生态位稳定性或适应性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


