Gene regulatory networks in disease and ageing
IF 28.6
1区 医学
Q1 UROLOGY & NEPHROLOGY
引用次数: 0
Abstract
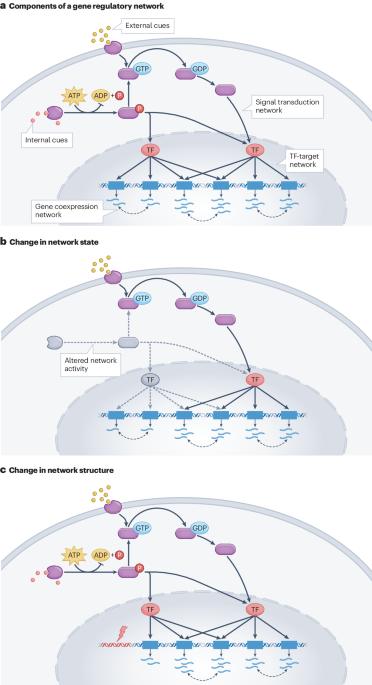
The precise control of gene expression is required for the maintenance of cellular homeostasis and proper cellular function, and the declining control of gene expression with age is considered a major contributor to age-associated changes in cellular physiology and disease. The coordination of gene expression can be represented through models of the molecular interactions that govern gene expression levels, so-called gene regulatory networks. Gene regulatory networks can represent interactions that occur through signal transduction, those that involve regulatory transcription factors, or statistical models of gene–gene relationships based on the premise that certain sets of genes tend to be coexpressed across a range of conditions and cell types. Advances in experimental and computational technologies have enabled the inference of these networks on an unprecedented scale and at unprecedented precision. Here, we delineate different types of gene regulatory networks and their cell-biological interpretation. We describe methods for inferring such networks from large-scale, multi-omics datasets and present applications that have aided our understanding of cellular ageing and disease mechanisms. Perturbations in the regulation of gene expression can contribute to disease- and ageing-associated changes in cell physiology. This review describes how the coordination of gene expression within and between cells can be represented through models of the molecular interactions that govern gene expression levels, and how such models can be used to understand age-associated changes in cell physiology.
疾病和老化中的基因调控网络
基因表达的精确控制是维持细胞稳态和细胞正常功能的必要条件,而随着年龄的增长,基因表达控制能力的下降被认为是造成与年龄相关的细胞生理学和疾病变化的主要原因。基因表达的协调可以通过控制基因表达水平的分子相互作用模型(即所谓的基因调控网络)来体现。基因调控网络可以代表通过信号转导发生的相互作用,也可以代表涉及调控转录因子的相互作用,还可以代表基因与基因关系的统计模型,其前提是某些基因集往往在一系列条件和细胞类型中共同表达。实验和计算技术的进步使这些网络的推断达到了前所未有的规模和精度。在这里,我们描述了不同类型的基因调控网络及其细胞生物学解释。我们介绍了从大规模多组学数据集中推断此类网络的方法,并介绍了有助于我们理解细胞老化和疾病机制的应用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Reviews Nephrology
医学-泌尿学与肾脏学
CiteScore
39.00
自引率
1.20%
发文量
127
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Nephrology aims to be the premier source of reviews and commentaries for the scientific communities it serves.
It strives to publish authoritative, accessible articles.
Articles are enhanced with clearly understandable figures, tables, and other display items.
Nature Reviews Nephrology publishes Research Highlights, News & Views, Comments, Reviews, Perspectives, and Consensus Statements.
The content is relevant to nephrologists and basic science researchers.
The broad scope of the journal ensures that the work reaches the widest possible audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


