Transcriptional regulators with broad expression in the zebrafish spinal cord
Abstract
Background
The spinal cord is a crucial part of the vertebrate CNS, controlling movements and receiving and processing sensory information from the trunk and limbs. However, there is much we do not know about how this essential organ develops. Here, we describe expression of 21 transcription factors and one transcriptional regulator in zebrafish spinal cord.
Results
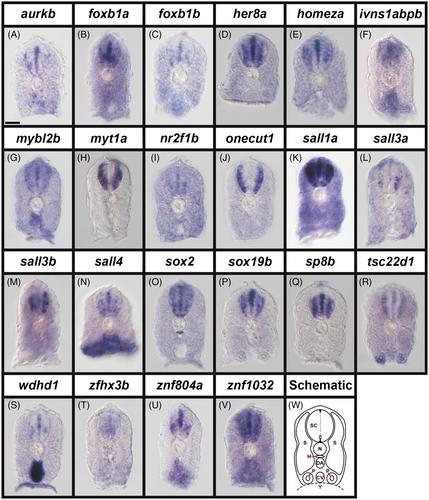
We analyzed the expression of aurkb, foxb1a, foxb1b, her8a, homeza, ivns1abpb, mybl2b, myt1a, nr2f1b, onecut1, sall1a, sall3a, sall3b, sall4, sox2, sox19b, sp8b, tsc22d1, wdhd1, zfhx3b, znf804a, and znf1032 in wild-type and MIB E3 ubiquitin protein ligase 1 zebrafish embryos. While all of these genes are broadly expressed in spinal cord, they have distinct expression patterns from one another. Some are predominantly expressed in progenitor domains, and others in subsets of post-mitotic cells. Given the conservation of spinal cord development, and the transcription factors and transcriptional regulators that orchestrate it, we expect that these genes will have similar spinal cord expression patterns in other vertebrates, including mammals and humans.
Conclusions
Our data identify 22 different transcriptional regulators that are strong candidates for playing different roles in spinal cord development. For several of these genes, this is the first published description of their spinal cord expression.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


