Innovative Self-Assembly of 15-Mer Chimeric α-Peptide–Oligourea Foldamers toward Cl−-Selective Ion Channels
IF 11.1
Q1 MATERIALS SCIENCE, MULTIDISCIPLINARY
引用次数: 0
Abstract
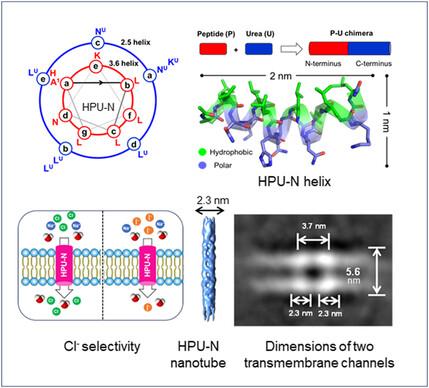
Constructing artificial ion channels is a challenging task. Herein, the de novo design of transmembrane ion channels made up of amphiphilic peptide–oligourea chimeric helices is described. They consist of an oligourea segment (7-mer) attached to the C-terminus of a short peptide (8-mer). Mass spectrometry (MS) and transmission electron microscopy (TEM) analyses show that in an aqueous solution, two of these chimeras (HPU-E and HPU-N) independently form defined oligomeric structures. TEM also shows that they form fiber bundles. The third related chimera HPU-F does not oligomerize (MS) but forms spherical nanostructures (TEM). HPU-E and HPU-N exhibit anion transport activity across lipid bilayers via antiport mechanism (HPU-N > HPU-E). The anion selectivity of HPU-N is Cl−>NO3− > Br−>SCN− > I− > AcO−>F−, which can be due to anion binding within the channels rather than size exclusion. Patch-clamp data support HPU-N's Cl− selectivity (PCl−/PI− = 3.26). X-ray crystal structure (1.77 Å) of HPU-N reveals well-packed α-helices, and cryo-electron microscopy data shows the formation of nanotubes (13.7 Å diameter pores) and transmembrane channels. The study shows that α-peptide–oligourea-based de novo design can yield unique bioactive molecules with defined structures and functions.
面向 Cl 选择性离子通道的 15 单体嵌合体 α 肽-寡糖脲折叠体的创新性自组装
构建人工离子通道是一项具有挑战性的任务。本文介绍了由两亲肽-寡脲嵌合螺旋组成的跨膜离子通道的全新设计。它们由连接到短肽(8-mer)C 端的低聚脲片段(7-mer)组成。质谱(MS)和透射电子显微镜(TEM)分析表明,在水溶液中,其中两种嵌合体(HPU-E 和 HPU-N)可独立形成确定的低聚结构。TEM 还显示它们形成了纤维束。第三个相关嵌合体 HPU-F 没有形成低聚物(MS),但形成了球形纳米结构(TEM)。HPU-E 和 HPU-N 通过反向传输机制(HPU-N > HPU-E)在脂质双分子层上表现出阴离子传输活性。HPU-N 的阴离子选择性为 Cl->NO3- >Br->SCN- >I- >AcO->F- ,这可能是由于阴离子在通道内结合而非尺寸排斥所致。膜片钳数据支持 HPU-N 的 Cl-选择性(PCl-/PI- = 3.26)。HPU-N 的 X 射线晶体结构(1.77 Å)显示了良好堆积的 α-螺旋,冷冻电镜数据显示了纳米管(直径 13.7 Å 的孔)和跨膜通道的形成。这项研究表明,基于α肽-寡糖脲的从头设计可以产生具有确定结构和功能的独特生物活性分子。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
14.00
自引率
2.40%
发文量
0
期刊介绍:
Small Science is a premium multidisciplinary open access journal dedicated to publishing impactful research from all areas of nanoscience and nanotechnology. It features interdisciplinary original research and focused review articles on relevant topics. The journal covers design, characterization, mechanism, technology, and application of micro-/nanoscale structures and systems in various fields including physics, chemistry, materials science, engineering, environmental science, life science, biology, and medicine. It welcomes innovative interdisciplinary research and its readership includes professionals from academia and industry in fields such as chemistry, physics, materials science, biology, engineering, and environmental and analytical science. Small Science is indexed and abstracted in CAS, DOAJ, Clarivate Analytics, ProQuest Central, Publicly Available Content Database, Science Database, SCOPUS, and Web of Science.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


