All-optical voltage imaging-guided postsynaptic single-cell transcriptome profiling with Voltage-Seq
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
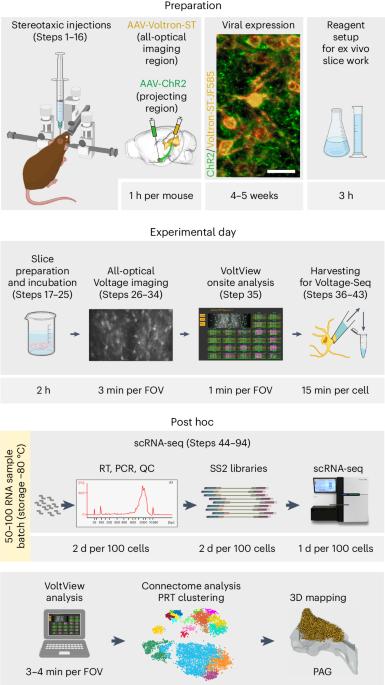
Neuronal pathways recruit large postsynaptic populations and maintain connections via distinct postsynaptic response types (PRTs). Until recently, PRTs were accessible as a selection criterion for single-cell RNA sequencing only through probing by low-throughput whole-cell electrophysiology. To overcome these limitations and target neurons on the basis of specific PRTs for soma collection and subsequent single-cell RNA sequencing, we developed Voltage-Seq using the genetically encoded voltage indicator Voltron in acute brain slices from mice. We also created an onsite analysis tool, VoltView, to guide soma collection of specific PRTs using a classifier based on a previously acquired database of connectomes from multiple animals. Here we present our procedure for preparing the optical path, the imaging setup and detailing the imaging and analysis steps, as well as a complete procedure for sequencing library preparation. This enables researchers to conduct our high-throughput all-optical synaptic assay and to obtain single-cell transcriptomic data from selected postsynaptic neurons. This also allows researchers to resolve the connectivity ratio of a specific pathway and explore the diversity of PRTs within that connectome. Furthermore, combining high throughput with quick analysis gives unique access to find specific connections within a large postsynaptic connectome. Voltage-Seq also allows the investigation of correlations between connectivity and gene expression changes in a postsynaptic cell-type-specific manner for both excitatory and inhibitory connections. The Voltage-Seq workflow can be completed in ~6 weeks, including 4–5 weeks for viral expression of the Voltron sensor. The technique requires knowledge of basic laboratory techniques, micromanipulator handling skills and experience in molecular biology and bioinformatics. Voltage-Seq is a method for all-optical voltage imaging-guided postsynaptic single-cell transcriptomics. It combines the use of the Voltron voltage indicator with the analysis tool VoltView to select specific neuronal somas to collect for single-cell RNA sequencing.
利用 Voltage-Seq 进行全光学电压成像引导的突触后单细胞转录组分析。
神经元通路通过不同的突触后反应类型(PRTs)招募大量突触后群体并维持连接。直到最近,PRTs 作为单细胞 RNA 测序的选择标准还只能通过低通量的全细胞电生理学探测来获得。为了克服这些限制,并根据特定的 PRTs 对神经元进行体节收集和随后的单细胞 RNA 测序,我们开发了 Voltage-Seq,在小鼠急性脑切片中使用基因编码的电压指示器 Voltron。我们还创建了一个现场分析工具 VoltView,利用基于先前获得的多动物连接组数据库的分类器指导特定 PRT 的体块采集。在这里,我们介绍了光路准备程序、成像设置、成像和分析步骤的细节,以及测序文库制备的完整程序。这样,研究人员就能进行高通量全光学突触测定,并从选定的突触后神经元获取单细胞转录组数据。这也使研究人员能够确定特定通路的连接比率,并探索该连接组中 PRT 的多样性。此外,将高通量与快速分析相结合,还能在庞大的突触后连接组中找到特定的连接。Voltage-Seq 还能以突触后细胞类型特异性的方式研究兴奋性和抑制性连接中连接性和基因表达变化之间的相关性。Voltage-Seq工作流程可在约6周内完成,其中包括4-5周的Voltron传感器病毒表达时间。这项技术要求具备基本的实验室技术知识、微机械手操作技能以及分子生物学和生物信息学方面的经验。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


