Real-time imaging of axonal membrane protein life cycles
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
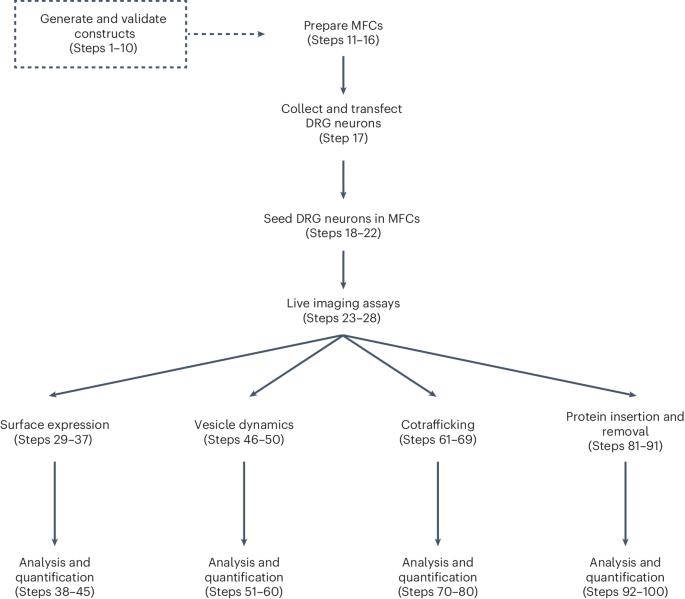
The construction of neuronal membranes is a dynamic process involving the biogenesis, vesicular packaging, transport, insertion and recycling of membrane proteins. Optical imaging is well suited for the study of protein spatial organization and transport. However, various shortcomings of existing imaging techniques have prevented the study of specific types of proteins and cellular processes. Here we describe strategies for protein tagging and labeling, cell culture and microscopy that enable the real-time imaging of axonal membrane protein trafficking and subcellular distribution as they progress through some stages of their life cycle. First, we describe a process for engineering membrane proteins with extracellular self-labeling tags (either HaloTag or SNAPTag), which can be labeled with fluorescent ligands of various colors and cell permeability, providing flexibility for investigating the trafficking and spatiotemporal regulation of multiple membrane proteins in neuronal compartments. Next, we detail the dissection, transfection and culture of dorsal root ganglion sensory neurons in microfluidic chambers, which physically compartmentalizes cell bodies and distal axons. Finally, we describe four labeling and imaging procedures that utilize these enzymatically tagged proteins, flexible fluorescent labels and compartmentalized neuronal cultures to study axonal membrane protein anterograde and retrograde transport, the cotransport of multiple proteins, protein subcellular localization, exocytosis and endocytosis. Additionally, we generated open-source software for analyzing the imaging data in a high throughput manner. The experimental and analysis workflows provide an approach for studying the dynamics of neuronal membrane protein homeostasis, addressing longstanding challenges in this area. The protocol requires 5–7 days and expertise in cell culture and microscopy. Conjugation of self-labeling enzymatic tags to axonal membrane proteins enables studying the dynamics of their trafficking, cellular localization and fate.
轴突膜蛋白生命周期的实时成像。
神经元膜的构建是一个动态过程,涉及膜蛋白的生物生成、囊泡包装、运输、插入和再循环。光学成像非常适合研究蛋白质的空间组织和运输。然而,现有成像技术的各种缺陷阻碍了对特定类型蛋白质和细胞过程的研究。在这里,我们介绍了蛋白质标记和标签、细胞培养和显微镜技术,这些技术可以对轴突膜蛋白在其生命周期的某些阶段的运输和亚细胞分布进行实时成像。首先,我们介绍了带有细胞外自标记标签(HaloTag 或 SNAPTag)的膜蛋白工程化过程,这些标签可以用不同颜色和细胞渗透性的荧光配体标记,为研究神经元区室中多种膜蛋白的贩运和时空调控提供了灵活性。接下来,我们将详细介绍背根神经节感觉神经元在微流体室中的解剖、转染和培养过程,该过程对细胞体和远端轴突进行了物理分隔。最后,我们介绍了四种标记和成像程序,这些程序利用这些酶标记蛋白、灵活的荧光标记和分室神经元培养来研究轴突膜蛋白的前向和逆向运输、多种蛋白的共运输、蛋白亚细胞定位、外吞和内吞。此外,我们还开发了开源软件,用于高通量分析成像数据。实验和分析工作流程提供了一种研究神经元膜蛋白动态平衡的方法,解决了这一领域长期存在的难题。该方案需要 5-7 天时间和细胞培养与显微镜方面的专业知识。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


