Quantifying Microsecond Solution-Phase Conformational Dynamics of a DNA Hairpin at the Single-Molecule Level
IF 3.7
Q2 CHEMISTRY, PHYSICAL
引用次数: 0
Abstract
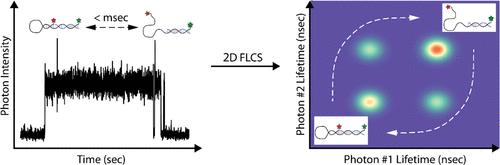
Quantifying the rapid conformational dynamics of biological systems is fundamental to understanding the mechanism. However, biomolecules are complex, often containing static and dynamic heterogeneity, thus motivating the use of single-molecule methods, particularly those that can operate in solution. In this study, we measure microsecond conformational dynamics of solution-phase DNA hairpins at the single-molecule level using an anti-Brownian electrokinetic (ABEL) trap. Different conformational states were distinguished by their fluorescence lifetimes, and kinetic parameters describing transitions between these states were determined using two-dimensional fluorescence lifetime correlation (2DFLCS) analysis. Rather than combining fluorescence signals from the entire data set ensemble, long observation times of individual molecules allowed ABEL-2DFLCS to be performed on each molecule independently, yielding the underlying distribution of the system’s kinetic parameters. ABEL-2DFLCS on the DNA hairpins resolved an underlying heterogeneity of fluorescence lifetimes and provided signatures of two-state exponential dynamics with rapid (<millisecond) transition times between states without observation of the substantially stretched exponential kinetics that had been observed in previous measurements on diffusing molecules. Numerical simulations were performed to validate the accuracy of this technique and the effects the underlying heterogeneity has on the analysis. Finally, ABEL-2DFLCS was performed on a mixture of hairpins and used to resolve their kinetic data.
在单分子水平量化 DNA 发夹的微秒级溶液相构象动力学
量化生物系统的快速构象动态是了解其机理的基础。然而,生物大分子非常复杂,通常包含静态和动态异质性,因此需要使用单分子方法,特别是那些可以在溶液中操作的方法。在这项研究中,我们利用反布朗电动(ABEL)陷阱在单分子水平上测量了溶液相 DNA 发夹的微秒构象动态。通过荧光寿命区分了不同的构象状态,并利用二维荧光寿命相关(2DFLCS)分析确定了描述这些状态之间转换的动力学参数。由于单个分子的观测时间较长,因此不需要合并整个数据集的荧光信号,而是对每个分子独立执行 ABEL-2DFLCS,从而得出系统动力学参数的基本分布。DNA 发夹的 ABEL-2DFLCS 解决了荧光寿命的潜在异质性,并提供了两态指数动力学的特征,状态之间的转换时间很快(毫秒级),而没有观察到以前对扩散分子进行测量时观察到的大幅拉伸的指数动力学。我们进行了数值模拟,以验证这一技术的准确性以及潜在的异质性对分析的影响。最后,对发夹混合物进行了 ABEL-2DFLCS 分析,并用于解析其动力学数据。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
3.70
自引率
0.00%
发文量
0
期刊介绍:
ACS Physical Chemistry Au is an open access journal which publishes original fundamental and applied research on all aspects of physical chemistry. The journal publishes new and original experimental computational and theoretical research of interest to physical chemists biophysical chemists chemical physicists physicists material scientists and engineers. An essential criterion for acceptance is that the manuscript provides new physical insight or develops new tools and methods of general interest. Some major topical areas include:Molecules Clusters and Aerosols; Biophysics Biomaterials Liquids and Soft Matter; Energy Materials and Catalysis

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


