Bartonella species in dromedaries and ruminants from Lower Shabelle and Benadir regions, Somalia
Abstract
Background
Bartonellosis, caused by bacteria of the genus Bartonella, is a zoonotic disease with several mammalian reservoir hosts. In Somalia, a country heavily reliant on livestock, zoonotic diseases pose significant public health and economic challenges. To the best of our knowledge, no study has been performed aiming to verify the occurrence of Bartonella spp. in Somalia. This study investigated the occurrence and molecular characterization of Bartonella in dromedary (Camelus dromedarius, Linnaeus, 1758), cattle, sheep, and goats from Somalia.
Materials and Methods
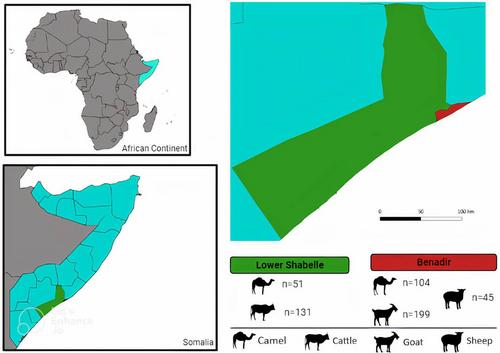
530 blood samples were collected from various animals (155 dromedary, 199 goat, 131 cattle, and 45 sheep) in Benadir and Lower Shabelle regions. DNA was extracted for molecular analysis, and a qPCR assay targeting the NADH dehydrogenase gamma subunit (nuoG) gene was used for Bartonella screening. Positive samples were also subjected to PCR assays targeting seven molecular markers including: nuoG, citrate synthase gene (gltA), RNA polymerase beta-subunit gene (rpoB), riboflavin synthase gene (ribC), 60 kDa heat-shock protein gene (groEL), cell division protein gene (ftsZ), and pap31 and qPCR targeting the 16-23S rRNA internal transcribed spacer (ITS) followed by Sanger sequencing, BLASTn and phylogenetic analysis.
Results
Out of 530 tested animals, 5.1% were positive for Bartonella spp. by the nuoG qPCR assay. Goats showed the highest Bartonella occurrence (17/199, 8.5%), followed by sheep (6/44, 6.8%), cattle (4/131, 3.1%), and dromedary (1/155, 1.9%). Goats, sheep, and cattle had higher odds of infection compared to dromedary. Among nuoG qPCR-positive samples, 11.1%, 14.8%, 11.1%, and 25.9% were positive in PCR assays based on nuoG, gltA, and pap31 genes, and in the qPCR based on the ITS region, respectively. On the other hand, nuoG qPCR-positive samples were negative in the PCR assays targeting the ribC, rpoB, ftsZ, and groEL genes. While Bartonella bovis sequences were detected in cattle (nuoG and ITS) and goats (gltA), Bartonella henselae ITS sequences were detected in dromedary, goat, and sheep. Phylogenetic analysis placed gltA Bartonella sequence from a goat in the same clade of B. bovis.
Conclusion
The present study showed, for the first time, molecular evidence of Bartonella spp. in dromedary and ruminants from Somalia and B. henselae in sheep and goats globally. These findings contribute valuable insights into Bartonella spp. occurrence in Somali livestock, highlighting the need for comprehensive surveillance and control measures under the One Health approach.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


