Cotton transposon-related variome reveals roles of transposon-related variations in modern cotton cultivation
IF 11.4
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
Introduction
Transposon plays a vital role in cotton genome evolution, contributing to the expansion and divergence of genomes within the Gossypium genus. However, knowledge of transposon activity in modern cotton cultivation is limited.
Objectives
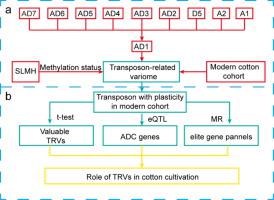
In this study, we aimed to construct transposon-related variome within Gossypium genus and reveal role of transposon-related variations during cotton cultivation. In addition, we try to identify valuable transposon-related variations for cotton breeding.
Methods
We utilized graphical genome construction to build up the graphical transposon-related variome. Based on the graphical variome, we integrated t-test, eQTL analysis and Mendelian Randomization (MR) to identify valuable transposon activities and elite genes. In addition, a convolutional neural network (CNN) model was constructed to evaluate epigenomic effects of transposon-related variations.
Results
We identified 35,980 transposon activities among 10 cotton genomes, and the diversity of genomic and epigenomic features was observed among 21 transposon categories. The graphical cotton transposon-related variome was constructed, and 9,614 transposon-related variations with plasticity in the modern cotton cohort were used for eQTL, phenotypic t-test and Mendelian Randomization. 128 genes were identified as gene resources improving fiber length and strength simultaneously. 4 genes were selected from 128 genes to construct the elite gene panel whose utility has been validated in a natural cotton cohort and 2 accessions with phenotypic divergence. Based on the eQTL analysis results, we identified transposon-related variations involved in cotton’s environmental adaption and human domestication, providing evidence of their role in cotton’s adaption-domestication cooperation.
Conclusions
The cotton transposon-related variome revealed the role of transposon-related variations in modern cotton cultivation, providing genomic resources for cotton molecular breeding.

棉花转座子相关变异组揭示了转座子相关变异在现代棉花种植中的作用。
导言:转座子在棉花基因组进化中发挥着重要作用,促进了棉属植物基因组的扩展和分化。然而,人们对现代棉花栽培中转座子活动的了解还很有限:本研究旨在构建棉花属中与转座子相关的变异组,揭示转座子相关变异在棉花栽培过程中的作用。此外,我们还试图找出对棉花育种有价值的转座子相关变异:方法:我们利用图形基因组构建技术建立了图形转座子相关变异组。在图形变异组的基础上,我们整合了t检验、eQTL分析和孟德尔随机化(Mendelian Randomization,MR),以鉴定有价值的转座子活动和精英基因。此外,我们还构建了一个卷积神经网络(CNN)模型,以评估转座子相关变异的表观基因组效应:结果:我们在 10 个棉花基因组中发现了 35,980 个转座子活动,在 21 个转座子类别中观察到了基因组和表观基因组特征的多样性。构建了图形化的棉花转座子相关变异组,并将现代棉花队列中具有可塑性的 9,614 个转座子相关变异用于 eQTL、表型 t 检验和孟德尔随机化。确定了 128 个基因作为同时改善纤维长度和强度的基因资源。从 128 个基因中筛选出 4 个基因,构建精英基因面板,其效用已在天然棉花群和 2 个表型差异较大的品种中得到验证。根据 eQTL 分析结果,我们确定了参与棉花环境适应和人类驯化的转座子相关变异,为其在棉花适应-驯化合作中的作用提供了证据:棉花转座子相关变异组揭示了转座子相关变异在现代棉花栽培中的作用,为棉花分子育种提供了基因组资源。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


