A new TGF-β risk score predicts clinical and immune landscape in colorectal cancer patients
Abstract
Background
Aberrant TGF-β signaling pathway can lead to invasive phenotype of colorectal cancer (CRC), resulting in poor prognosis. It is pivotal to develop an effective prognostic factor on the basis of TGF-β-related genes to accurately identify risk of CRC patients.
Methods
We performed differential analysis of TGF-β-related genes in CRC patients from databases and previous literature to obtain TGF-β-related differentially expressed genes (TRDEGs). LASSO-Cox regression was utilized to build a CRC prognostic feature model based on TRDEGs. The model was validated using two GEO validation sets. Wilcoxon rank-sum test was utilized to test correlation of model with clinical factors. ESTIMATE algorithm and ssGSEA and tumor mutation burden (TMB) analysis were used to analyze immune landscape and mutation burden of high-risk (HR) and low-risk (LR) groups. CellMiner database was utilized to identify therapeutic drugs with high sensitivity to the feature genes.
Results
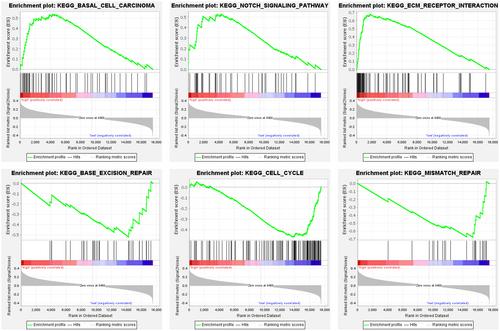
We established a six-gene risk prognostic model with good predictive accuracy, which independently predicted CRC patients' prognoses. The HR group was more likely to experience immunotherapy benefits due to higher immune infiltration and TMB. The feature gene TGFB2 could inhibit the efficacy of drugs such as XAV-939, Staurosporine, and Dasatinib, but promote the efficacy of drugs such as CUDC-305 and by-product of CUDC-305. Similarly, RBL1 could inhibit the drug action of Fluphenazine and Imiquimod but promote that of Irofulven.
Conclusion
A CRC risk prognostic signature was developed on basis of TGF-β-related genes, which provides a reference for risk and further therapeutic selection of CRC patients.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


