Structural insights into PPP2R5A degradation by HIV-1 Vif
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
HIV-1 Vif recruits host cullin-RING-E3 ubiquitin ligase and CBFβ to degrade the cellular APOBEC3 antiviral proteins through diverse interactions. Recent evidence has shown that Vif also degrades the regulatory subunits PPP2R5(A–E) of cellular protein phosphatase 2A to induce G2/M cell cycle arrest. As PPP2R5 proteins bear no functional or structural resemblance to A3s, it is unclear how Vif can recognize different sets of proteins. Here we report the cryogenic-electron microscopy structure of PPP2R5A in complex with HIV-1 Vif–CBFβ–elongin B–elongin C at 3.58 Å resolution. The structure shows PPP2R5A binds across the Vif molecule, with biochemical and cellular studies confirming a distinct Vif–PPP2R5A interface that partially overlaps with those for A3s. Vif also blocks a canonical PPP2R5A substrate-binding site, indicating that it suppresses the phosphatase activities through both degradation-dependent and degradation-independent mechanisms. Our work identifies critical Vif motifs regulating the recognition of diverse A3 and PPP2R5A substrates, whereby disruption of these host–virus protein interactions could serve as potential targets for HIV-1 therapeutics. The authors solve a cryo-EM structure of the regulatory subunit of human protein phosphatase 2A in complex with HIV-1 Vif-containing E3 ligase, leading to improvement of our understanding of host–virus protein interactions.

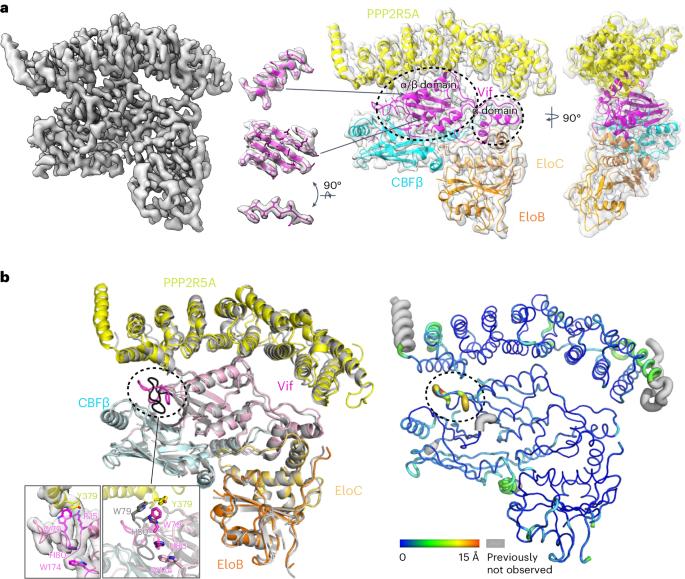
艾滋病毒-1 Vif 对 PPP2R5A 降解的结构性启示
HIV-1 Vif通过多种相互作用,招募宿主cullin-RING-E3泛素连接酶和CBFβ降解细胞APOBEC3抗病毒蛋白。最近的证据表明,Vif 还能降解细胞蛋白磷酸酶 2A 的调节亚基 PPP2R5(A-E),从而诱导 G2/M 细胞周期停滞。由于 PPP2R5 蛋白在功能或结构上与 A3s 没有相似之处,因此 Vif 如何识别不同的蛋白质尚不清楚。在此,我们以 3.58 Å 的分辨率报告了 PPP2R5A 与 HIV-1 Vif-CBFβ-elongin B-elongin C 复合物的低温电子显微镜结构。该结构显示 PPP2R5A 跨 Vif 分子结合,生化和细胞研究证实 Vif-PPP2R5A 界面与 A3 界面部分重叠。Vif 还阻断了一个典型的 PPP2R5A 底物结合位点,表明它通过依赖降解和不依赖降解两种机制抑制磷酸酶的活性。我们的研究发现了调节对不同 A3 和 PPP2R5A 底物识别的关键 Vif 基序,因此破坏这些宿主-病毒蛋白的相互作用可作为 HIV-1 疗法的潜在靶点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


