ProBac-seq, a bacterial single-cell RNA sequencing methodology using droplet microfluidics and large oligonucleotide probe sets
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Methods that measure the transcriptomic state of thousands of individual cells have transformed our understanding of cellular heterogeneity in eukaryotic cells since their introduction in the past decade. While simple and accessible protocols and commercial products are now available for the processing of mammalian cells, these existing technologies are incompatible with use in bacterial samples for several fundamental reasons including the absence of polyadenylation on bacterial messenger RNA, the instability of bacterial transcripts and the incompatibility of bacterial cell morphology with existing methodologies. Recently, we developed ProBac sequencing (ProBac-seq), a method that overcomes these technical difficulties and provides high-quality single-cell gene expression data from thousands of bacterial cells by using messenger RNA-specific probes. Here we provide details for designing large oligonucleotide probe sets for an organism of choice, amplifying probe sets to produce sufficient quantities for repeated experiments, adding unique molecular indexes and poly-A tails to produce finalized probes, in situ probe hybridization and single-cell encapsulation and library preparation. This protocol, from the probe amplification to the library preparation, requires ~7 d to complete. ProBac-seq offers several advantages over other methods by capturing only the desired target sequences and avoiding nondesired transcripts, such as highly abundant ribosomal RNA, thus enriching for signal that better informs on cellular state. The use of multiple probes per gene can detect meaningful single-cell signals from cells expressing transcripts to a lesser degree or those grown in minimal media and other environmentally relevant conditions in which cells are less active. ProBac-seq is also compatible with other organisms that can be profiled by in situ hybridization techniques. This protocol presents ProBac sequencing, a bacterial single-cell RNA sequencing methodology using droplet microfluidics and large oligonucleotide probe sets.
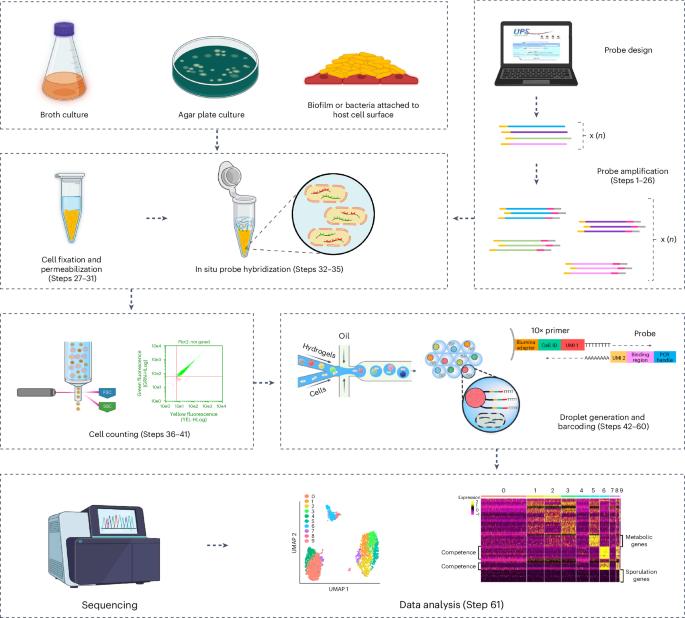
ProBac-seq 是一种使用液滴微流控技术和大型寡核苷酸探针组的细菌单细胞 RNA 测序方法。
测量数千个单个细胞转录组状态的方法自过去十年问世以来,改变了我们对真核细胞异质性的理解。虽然目前已有用于处理哺乳动物细胞的简单易行的方案和商业产品,但由于细菌信使 RNA 没有多聚腺苷酸化、细菌转录本不稳定以及细菌细胞形态与现有方法不兼容等几个根本原因,这些现有技术无法用于细菌样本。最近,我们开发了 ProBac 测序(ProBac-seq),这种方法克服了这些技术难题,通过使用信使 RNA 特异性探针从成千上万个细菌细胞中提供高质量的单细胞基因表达数据。在此,我们详细介绍了如何为所选生物设计大型寡核苷酸探针集、扩增探针集以产生足够数量的探针用于重复实验、添加独特的分子索引和聚 A 尾以产生最终探针、原位探针杂交以及单细胞封装和文库制备。该方案从探针扩增到文库制备,需要约 7 天才能完成。与其他方法相比,ProBac-seq 有几个优势:只捕获所需的目标序列,避免非所需的转录本,如高度丰富的核糖体 RNA,从而富集信号,更好地了解细胞状态。每个基因使用多个探针,可以从表达转录本程度较低的细胞或在最小培养基和其他环境相关条件下生长的细胞中检测到有意义的单细胞信号,因为在这些条件下细胞的活性较低。ProBac-seq 还与其他可通过原位杂交技术进行分析的生物体兼容。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


