A lactate-responsive gene signature predicts the prognosis and immunotherapeutic response of patients with triple-negative breast cancer
Abstract
Background
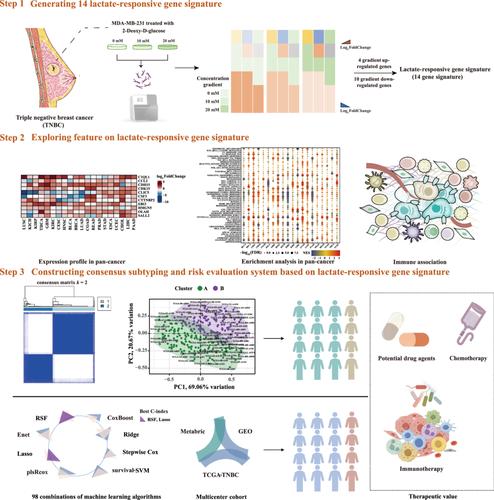
Increased glycolytic activity and lactate production are characteristic features of triple-negative breast cancer (TNBC). The aim of this study was to determine whether a subset of lactate-responsive genes (LRGs) could be used to classify TNBC subtypes and predict patient outcomes.
Methods
Lactate levels were initially measured in different breast cancer (BC) cell types. Subsequently, MDA-MB-231 cells treated with 2-Deoxy-d-glucose or l-lactate were subjected to RNA sequencing (RNA-seq). The gene set variation analysis algorithm was utilized to calculate the lactate-responsive score, conduct a differential analysis, and establish an association with the extent of immune infiltration. Consensus clustering was then employed to classify TNBC patients. Tumor immune dysfunction and exclusion, cibersort, single-sample gene set enrichment analysis, and EPIC, were used to compare the tumor-infiltrating immune cells between TNBC subtypes and predict the response to immunotherapy. Furthermore, a prognostic model was developed by combining 98 machine learning algorithms, to assess the predictive significance of the LRG signature. The predictive value of immune infiltration and the immunotherapy response was also assessed. Finally, the association between lactate and various anticancer drugs was examined based on expression profile similarity principles.
Results
We found that the lactate levels of TNBC cells were significantly higher than those of other BC cell lines. Through RNA-seq, we identified 14 differentially expressed LRGs in TNBC cells under varying lactate levels. Notably, this LRG signature was associated with interleukin-17 signaling pathway dysregulation, suggesting a link between lactate metabolism and immune impairment. Furthermore, the LRG signature was used to categorize TNBC into two distinct subtypes, whereby Subtype A was characterized by immunosuppression, whereas Subtype B was characterized by immune activation.
Conclusion
We identified an LRG signature in TNBC, which could be used to predict the prognosis of patients with TNBC and gauge their response to immunotherapy. Our findings may help guide the precision treatment of patients with TNBC.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


