The LexA–RecA* structure reveals a cryptic lock-and-key mechanism for SOS activation
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
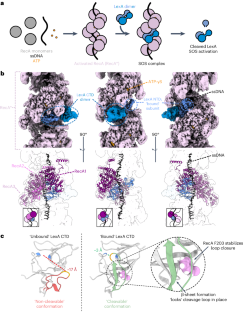
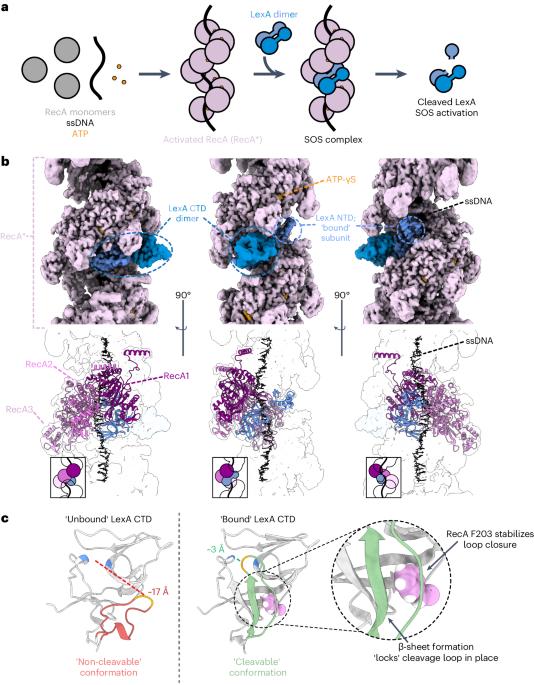
The bacterial SOS response plays a key role in adaptation to DNA damage, including genomic stress caused by antibiotics. SOS induction begins when activated RecA*, an oligomeric nucleoprotein filament that forms on single-stranded DNA, binds to and stimulates autoproteolysis of the repressor LexA. Here, we present the structure of the complete Escherichia coli SOS signal complex, constituting full-length LexA bound to RecA*. We uncover an extensive interface unexpectedly including the LexA DNA-binding domain, providing a new molecular rationale for ordered SOS gene induction. We further find that the interface involves three RecA subunits, with a single residue in the central engaged subunit acting as a molecular key, inserting into an allosteric binding pocket to induce LexA cleavage. Given the pro-mutagenic nature of SOS activation, our structural and mechanistic insights provide a foundation for developing new therapeutics to slow the evolution of antibiotic resistance. Here, using cryo-EM, the authors reveal the mechanism by which RecA filamented on single-stranded DNA binds to and induces LexA cleavage, the key signal governing the bacterial DNA damage response pathway implicated in antibiotic resistance.
LexA-RecA* 结构揭示了 SOS 激活的隐秘锁钥机制
细菌的 SOS 反应在适应 DNA 损伤(包括抗生素引起的基因组压力)方面发挥着关键作用。当活化的 RecA*(一种在单链 DNA 上形成的寡聚核蛋白丝)与抑制因子 LexA 结合并刺激其自体蛋白水解时,SOS 诱导就开始了。在这里,我们展示了完整的大肠杆菌 SOS 信号复合体结构,它由与 RecA* 结合的全长 LexA 构成。我们意外地发现了一个包括 LexA DNA 结合域在内的广泛界面,为有序的 SOS 基因诱导提供了新的分子原理。我们进一步发现,该界面涉及三个 RecA 亚基,中央参与亚基中的一个残基充当了分子钥匙,插入异生结合口袋,诱导 LexA 分裂。鉴于 SOS 激活具有促突变的性质,我们在结构和机理方面的见解为开发减缓抗生素耐药性演变的新疗法奠定了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


