Combating Prozone Effects and Predicting the Dynamic Range of Naked-Eye Nanoplasmonic Biosensors through Capture Bioentity Optimization
IF 4.6
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
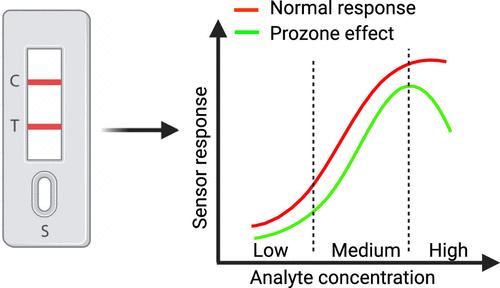
Accurately quantifying high analyte concentrations poses a challenge due to the common occurrence of the prozone or hook effect within sandwich assays utilized in plasmonic nanoparticle-based lateral flow devices (LFDs). As a result, LFDs are often underestimated compared to other biosensors with concerns surrounding their specificity and sensitivity toward the target analyte. To address this limitation, here we develop an analytical model capable of predicting the prozone effect and subsequently the dynamic range of the biosensor based on the concentration of the capture antibody. To support our model, we conduct a sandwich immunoassay to detect C-reactive protein (CRP) in a phosphate-buffered saline (PBS) buffer using an LFD. Within the experiment, we investigate the relationship between the CRP dynamic range and the prozone effect as a function of the capture antibody concentration, which is increased from 0.1 to 2 mg/mL. The experimental results, while supporting the developed analytical model, show that increasing the capture antibody concentration increases the dynamic range. The developed model therefore holds the potential to expand the measurable range and reduce costs associated with quantifying biomarkers in diverse diagnostic assays. This will ultimately allow LFDs to have better clinical significance before the prozone effect becomes dominant.
通过捕获生物特性优化消除原区效应并预测裸眼纳米光电生物传感器的动态范围
由于在基于等离子纳米粒子的横向流动装置(LFD)中使用的夹心检测法中经常出现前区或钩状效应,因此准确量化高浓度分析物是一项挑战。因此,与其他生物传感器相比,LFD 经常被低估,人们担心其对目标分析物的特异性和灵敏度。为了解决这一局限性,我们在此建立了一个分析模型,该模型能够根据捕获抗体的浓度预测原区效应以及生物传感器的动态范围。为了支持我们的模型,我们使用 LFD 进行了夹心免疫测定,以检测磷酸盐缓冲盐水 (PBS) 缓冲液中的 C 反应蛋白 (CRP)。在实验中,我们研究了 CRP 动态范围和原区效应与捕获抗体浓度(从 0.1 毫克/毫升增加到 2 毫克/毫升)之间的关系。实验结果支持所开发的分析模型,同时表明提高捕获抗体浓度会增加动态范围。因此,所开发的模型有可能扩大可测量范围,降低在各种诊断检测中量化生物标记物的相关成本。这最终将使 LFD 在原区效应占主导地位之前具有更好的临床意义。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Measurement Science Au
化学计量学-
CiteScore
5.20
自引率
0.00%
发文量
0
期刊介绍:
ACS Measurement Science Au is an open access journal that publishes experimental computational or theoretical research in all areas of chemical measurement science. Short letters comprehensive articles reviews and perspectives are welcome on topics that report on any phase of analytical operations including sampling measurement and data analysis. This includes:Chemical Reactions and SelectivityChemometrics and Data ProcessingElectrochemistryElemental and Molecular CharacterizationImagingInstrumentationMass SpectrometryMicroscale and Nanoscale systemsOmics (Genomics Proteomics Metabonomics Metabolomics and Bioinformatics)Sensors and Sensing (Biosensors Chemical Sensors Gas Sensors Intracellular Sensors Single-Molecule Sensors Cell Chips Arrays Microfluidic Devices)SeparationsSpectroscopySurface analysisPapers dealing with established methods need to offer a significantly improved original application of the method.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


