A spatial transcriptome map of the developing maize ear
IF 15.8
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
Abstract
A comprehensive understanding of inflorescence development is crucial for crop genetic improvement, as inflorescence meristems give rise to reproductive organs and determine grain yield. However, dissecting inflorescence development at the cellular level has been challenging owing to a lack of specific marker genes to distinguish among cell types, particularly in different types of meristems that are vital for organ formation. In this study, we used spatial enhanced resolution omics-sequencing (Stereo-seq) to construct a precise spatial transcriptome map of the developing maize ear primordium, identifying 12 cell types, including 4 newly defined cell types found mainly in the inflorescence meristem. By extracting the meristem components for detailed clustering, we identified three subtypes of meristem and validated two MADS-box genes that were specifically expressed at the apex of determinate meristems and involved in stem cell determinacy. Furthermore, by integrating single-cell RNA transcriptomes, we identified a series of spatially specific networks and hub genes that may provide new insights into the formation of different tissues. In summary, this study provides a valuable resource for research on cereal inflorescence development, offering new clues for yield improvement. The authors integrate spatial (Stereo-seq) and single-cell transcriptomes of the developing maize ear to produce an atlas of maize ear cells and their developmental trajectories. They also identify a pair of transcription factors involved in inflorescence development.
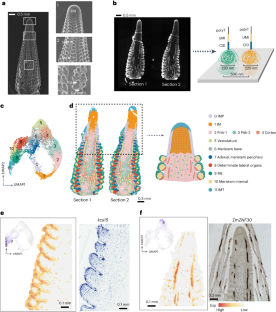
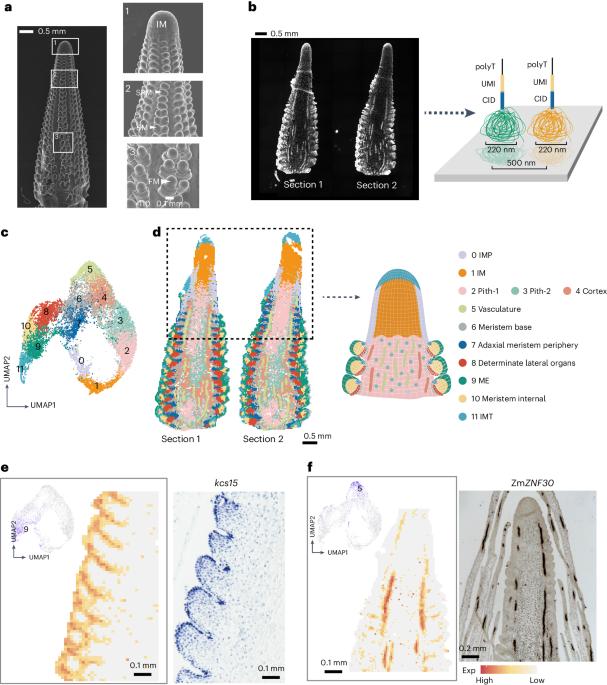
发育中玉米穗的空间转录组图谱
全面了解花序发育对作物遗传改良至关重要,因为花序分生组织产生生殖器官并决定谷物产量。然而,由于缺乏特异性标记基因来区分细胞类型,特别是对器官形成至关重要的不同类型的分生组织,因此在细胞水平上剖析花序发育一直是个挑战。在这项研究中,我们利用空间增强分辨率omics测序(Stereo-seq)构建了发育中玉米穗原基的精确空间转录组图谱,确定了12种细胞类型,包括主要存在于花序分生组织中的4种新定义的细胞类型。通过提取分生组织成分进行详细聚类,我们确定了分生组织的三个亚型,并验证了两个 MADS-box 基因,这两个基因在确定性分生组织的顶端特异表达,并参与干细胞的确定性。此外,通过整合单细胞 RNA 转录组,我们发现了一系列空间特异性网络和枢纽基因,它们可能为不同组织的形成提供新的见解。总之,这项研究为谷物花序发育研究提供了宝贵的资源,为提高产量提供了新的线索。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
文献相关原料
| 公司名称 | 产品信息 | 采购帮参考价格 |
|---|
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


