Novel progressive deep learning algorithm for uncovering multiple single nucleotide polymorphism interactions to predict paclitaxel clearance in patients with nonsmall cell lung cancer
Abstract
Background
The rate at which the anticancer drug paclitaxel is cleared from the body markedly impacts its dosage and chemotherapy effectiveness. Importantly, paclitaxel clearance varies among individuals, primarily because of genetic polymorphisms. This metabolic variability arises from a nonlinear process that is influenced by multiple single nucleotide polymorphisms (SNPs). Conventional bioinformatics methods struggle to accurately analyze this complex process and, currently, there is no established efficient algorithm for investigating SNP interactions.
Methods
We developed a novel machine-learning approach called GEP-CSIs data mining algorithm. This algorithm, an advanced version of GEP, uses linear algebra computations to handle discrete variables. The GEP-CSI algorithm calculates a fitness function score based on paclitaxel clearance data and genetic polymorphisms in patients with nonsmall cell lung cancer. The data were divided into a primary set and a validation set for the analysis.
Results
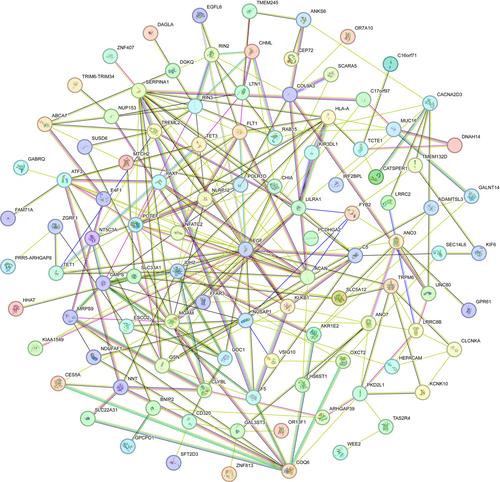
We identified and validated 1184 three-SNP combinations that had the highest fitness function values. Notably, SERPINA1, ATF3 and EGF were found to indirectly influence paclitaxel clearance by coordinating the activity of genes previously reported to be significant in paclitaxel clearance. Particularly intriguing was the discovery of a combination of three SNPs in genes FLT1, EGF and MUC16. These SNPs-related proteins were confirmed to interact with each other in the protein–protein interaction network, which formed the basis for further exploration of their functional roles and mechanisms.
Conclusion
We successfully developed an effective deep-learning algorithm tailored for the nuanced mining of SNP interactions, leveraging data on paclitaxel clearance and individual genetic polymorphisms.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


