Cryo-EM structure of the Rev1–Polζ holocomplex reveals the mechanism of their cooperativity in translesion DNA synthesis
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Rev1–Polζ-dependent translesion synthesis (TLS) of DNA is crucial for maintaining genome integrity. To elucidate the mechanism by which the two polymerases cooperate in TLS, we determined the cryogenic electron microscopic structure of the Saccharomyces cerevisiae Rev1–Polζ holocomplex in the act of DNA synthesis (3.53 Å). We discovered that a composite N-helix-BRCT module in Rev1 is the keystone of Rev1–Polζ cooperativity, interacting directly with the DNA template–primer and with the Rev3 catalytic subunit of Polζ. The module is positioned akin to the polymerase-associated domain in Y-family TLS polymerases and is set ideally to interact with PCNA. We delineate the full extent of interactions that the carboxy-terminal domain of Rev1 makes with Polζ and identify potential new druggable sites to suppress chemoresistance from first-line chemotherapeutics. Collectively, our results provide fundamental new insights into the mechanism of cooperativity between Rev1 and Polζ in TLS. The authors elucidate by cryo-EM the mechanism by which DNA polymerases Rev1 and Polζ cooperate in translesion DNA synthesis.
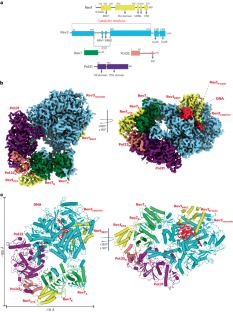
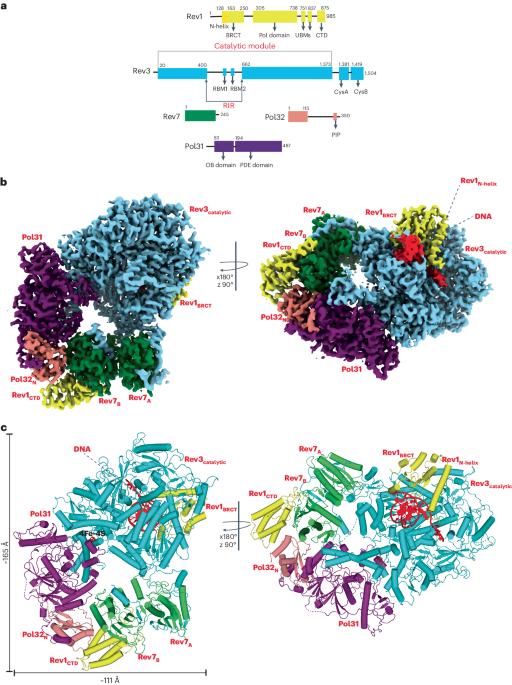
Rev1-Polζ 整体复合物的低温电子显微镜结构揭示了它们在转座子 DNA 合成中的合作机制
依赖于Rev1-Polζ的DNA转座合成(TLS)对于维持基因组的完整性至关重要。为了阐明这两种聚合酶在 TLS 中的合作机制,我们测定了正在进行 DNA 合成的酿酒酵母 Rev1-Polζ 整体复合体的低温电子显微镜结构(3.53 Å)。我们发现,Rev1 中的一个复合 N-helix-BRCT 模块是 Rev1-Polζ 协同作用的基石,它直接与 DNA 模板二聚体和 Polζ 的 Rev3 催化亚基相互作用。该模块的位置类似于Y-家族TLS聚合酶中的聚合酶相关结构域,是与PCNA相互作用的理想设置。我们描述了Rev1的羧基末端结构域与Polζ相互作用的全部过程,并确定了抑制一线化疗药物化疗耐药性的潜在新药物作用位点。总之,我们的研究结果为了解 Rev1 与 Polζ 在 TLS 中的合作机制提供了新的基本见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


