Cerebral organoids display dynamic clonal growth and tunable tissue replenishment
IF 17.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
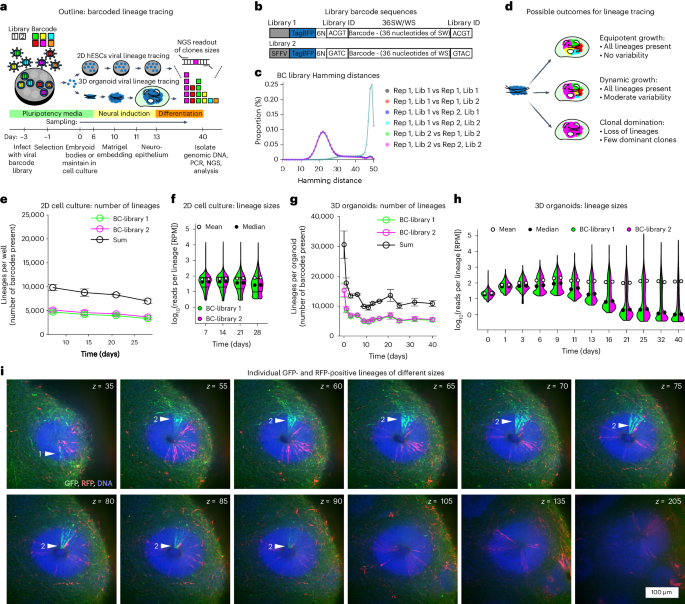
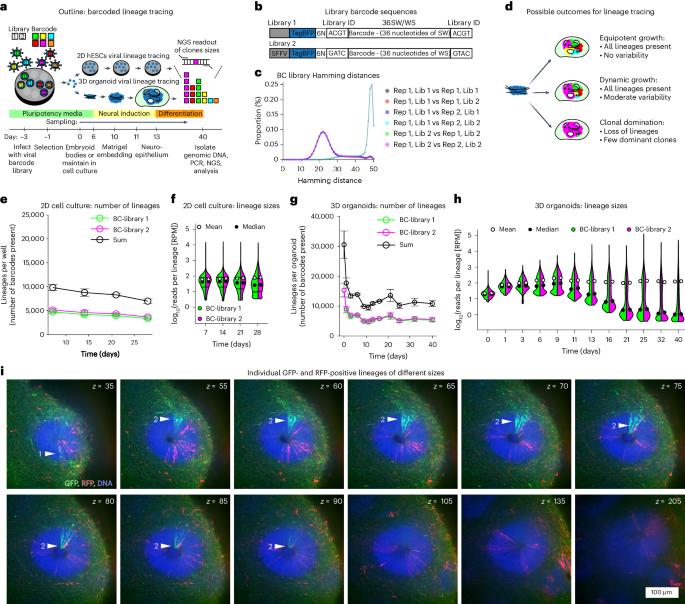
During brain development, neural progenitors expand through symmetric divisions before giving rise to differentiating cell types via asymmetric divisions. Transition between those modes varies among individual neural stem cells, resulting in clones of different sizes. Imaging-based lineage tracing allows for lineage analysis at high cellular resolution but systematic approaches to analyse clonal behaviour of entire tissues are currently lacking. Here we implement whole-tissue lineage tracing by genomic DNA barcoding in 3D human cerebral organoids, to show that individual stem cell clones produce progeny on a vastly variable scale. By using stochastic modelling we find that variable lineage sizes arise because a subpopulation of lineages retains symmetrically dividing cells. We show that lineage sizes can adjust to tissue demands after growth perturbation via chemical ablation or genetic restriction of a subset of cells in chimeric organoids. Our data suggest that adaptive plasticity of stem cell populations ensures robustness of development in human brain organoids. Lindenhofer, Haendeler, Esk, Littleboy et al. perform whole-tissue lineage tracing in human cerebral organoids to reveal that a subpopulation of symmetrically dividing cells can adjust its lineage size depending on tissue demands.
脑器官组织显示出动态克隆生长和可调的组织补充能力
在大脑发育过程中,神经祖细胞通过对称分裂扩大,然后通过不对称分裂产生分化细胞类型。不同的神经干细胞在这些模式之间的转换各不相同,从而形成不同大小的克隆。基于成像的系谱追踪可进行高细胞分辨率的系谱分析,但目前还缺乏分析整个组织克隆行为的系统方法。在这里,我们在三维人脑器官组织中通过基因组DNA条形码实现了全组织系谱追踪,显示单个干细胞克隆产生的后代规模变化很大。通过随机建模,我们发现不同的系大小是由于系的亚群保留了对称分裂的细胞。我们的研究表明,通过化学消融或基因限制嵌合有机体中的一个细胞亚群,在生长受到干扰后,细胞系的大小可以根据组织需求进行调整。我们的数据表明,干细胞群的适应性可塑性确保了人脑器官组织发育的稳健性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
文献相关原料
| 公司名称 | 产品信息 | 采购帮参考价格 |
|---|
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


