Three-dimensional dipole orientation mapping with high temporal-spatial resolution using polarization modulation
IF 15.7
Q1 OPTICS
引用次数: 0
Abstract
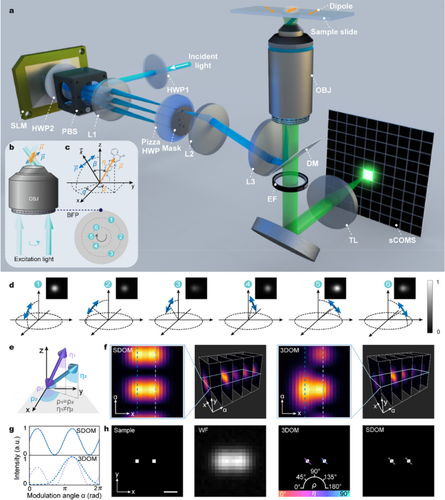
Fluorescence polarization microscopy is widely used in biology for molecular orientation properties. However, due to the limited temporal resolution of single-molecule orientation localization microscopy and the limited orientation dimension of polarization modulation techniques, achieving simultaneous high temporal-spatial resolution mapping of the three-dimensional (3D) orientation of fluorescent dipoles remains an outstanding problem. Here, we present a super-resolution 3D orientation mapping (3DOM) microscope that resolves 3D orientation by extracting phase information of the six polarization modulation components in reciprocal space. 3DOM achieves an azimuthal precision of 2° and a polar precision of 3° with spatial resolution of up to 128 nm in the experiments. We validate that 3DOM not only reveals the heterogeneity of the milk fat globule membrane, but also elucidates the 3D structure of biological filaments, including the 3D spatial conformation of λ-DNA and the structural disorder of actin filaments. Furthermore, 3DOM images the dipole dynamics of microtubules labeled with green fluorescent protein in live U2OS cells, reporting dynamic 3D orientation variations. Given its easy integration into existing wide-field microscopes, we expect the 3DOM microscope to provide a multi-view versatile strategy for investigating molecular structure and dynamics in biological macromolecules across multiple spatial and temporal scales.
利用偏振调制实现高时空分辨率的三维偶极子方位制图
荧光偏振显微镜在生物学中被广泛应用于分子定向特性的研究。然而,由于单分子取向定位显微镜的时间分辨率有限,偏振调制技术的取向维度也有限,因此实现荧光偶极子三维(3D)取向的同步高时空分辨率绘图仍是一个突出问题。在这里,我们提出了一种超分辨率三维方位映射(3DOM)显微镜,它通过提取倒易空间中六个偏振调制分量的相位信息来解析三维方位。在实验中,3DOM 实现了 2° 的方位精度和 3° 的极性精度,空间分辨率高达 128 nm。我们验证了3DOM不仅能揭示牛奶脂肪球膜的异质性,还能阐明生物丝的三维结构,包括λ-DNA的三维空间构象和肌动蛋白丝的结构紊乱。此外,3DOM 还能对活体 U2OS 细胞中用绿色荧光蛋白标记的微管的偶极动态进行成像,报告动态三维方向变化。鉴于3DOM显微镜易于集成到现有的宽视场显微镜中,我们希望它能为研究生物大分子的分子结构和动力学提供一种跨时空尺度的多视角多功能策略。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


