Decoding and reconstructing disease relations between dry eye and depression: a multimodal investigation comprising meta-analysis, genetic pathways and Mendelian randomization
IF 11.4
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
Introduction
The clinical presentations of dry eye disease (DED) and depression (DEP) often comanifest. However, the robustness and the mechanisms underlying this association were undetermined.
Objectives
To this end, we set up a three-segment study that employed multimodality results (meta-analysis, genome-wide association study [GWAS] and Mendelian randomization [MR]) to elucidate the association, common pathways and causality between DED and DEP.
Methods
A meta-analysis comprising 26 case−control studies was first conducted to confirm the DED-DEP association. Next, we performed a linkage disequilibrium (LD)-adjusted GWAS and targeted phenotype association study (PheWAS) in East Asian TW Biobank (TWB) and European UK Biobank (UKB) populations. Single-nucleotide polymorphisms (SNPs) were further screened for molecular interactions and common pathways at the functional gene level. To further elucidate the activated pathways in DED and DEP, a systemic transcriptome review was conducted on RNA sequencing samples from the Gene Expression Omnibus. Finally, 48 MR experiments were implemented to examine the bidirectional causation between DED and DEP.
Results
Our meta-analysis showed that DED patients are associated with an increased DEP prevalence (OR = 1.83), while DEP patients have a concurrent higher risk of DED (OR = 2.34). Notably, cross-disease GWAS analysis revealed that similar genetic architecture (rG = 0.19) and pleiotropic functional genes contributed to phenotypes in both diseases. Through protein−protein interaction and ontology convergence, we summarized the pleiotropic functional genes under the ontology of immune activation, which was further validated by a transcriptome systemic review. Importantly, the inverse variance-weighted (IVW)-MR experiments in both TWB and UKB populations (p value <0.001) supported the bidirectional exposure-outcome causation for DED-to-DEP and DEP-to-DED. Despite stringent LD-corrected instrumental variable re-selection, the bidirectional causation between DED and DEP remained.
Conclusion
With the multi-modal evidence combined, we consolidated the association and causation between DED and DEP.
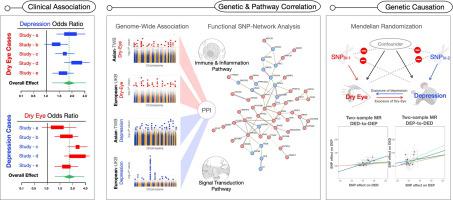

解码和重建干眼症与抑郁症之间的疾病关系:由 Meta 分析、遗传途径和孟德尔随机化组成的多模式研究。
导言:干眼症(DED)和抑郁症(DEP)的临床表现经常同时出现。然而,这种关联的稳健性及其机制尚未确定:为此,我们建立了一项三段式研究,采用多模态结果(荟萃分析、全基因组关联研究[GWAS]和孟德尔随机分析[MR])来阐明 DED 和 DEP 之间的关联、共同途径和因果关系:方法: 我们首先对 26 项病例对照研究进行了荟萃分析,以确认 DED 与 DEP 的关联。接下来,我们在东亚 TW Biobank(TWB)和欧洲 UK Biobank(UKB)人群中进行了经连接不平衡(LD)调整的 GWAS 和目标表型关联研究(PheWAS)。进一步筛选了单核苷酸多态性(SNPs),以确定功能基因水平上的分子相互作用和共同通路。为进一步阐明 DED 和 DEP 的激活通路,对基因表达总库(Gene Expression Omnibus)中的 RNA 测序样本进行了系统转录组审查。最后,我们进行了 48 项磁共振实验,以研究 DED 和 DEP 之间的双向因果关系:我们的荟萃分析表明,DED 患者与 DEP 患病率增加有关(OR = 1.83),而 DEP 患者同时具有较高的 DED 风险(OR = 2.34)。值得注意的是,跨疾病 GWAS 分析显示,相似的遗传结构(rG = 0.19)和多效功能基因对两种疾病的表型都有影响。通过蛋白质-蛋白质相互作用和本体汇聚,我们总结了免疫激活本体下的多效功能基因,并通过转录组系统回顾进一步验证了这一点。重要的是,反方差加权(IVW)-MR 在 TWB 和 UKB 群体中的实验结果(P 值 结论)表明,TWB 和 UKB 群体的免疫激活功能基因的 "多向性 "和 "多向性 "均高于 TWB 和 UKB:结合多种模式的证据,我们整合了 DED 和 DEP 之间的关联和因果关系。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Advanced Research
Multidisciplinary-Multidisciplinary
CiteScore
21.60
自引率
0.90%
发文量
280
审稿时长
12 weeks
期刊介绍:
Journal of Advanced Research (J. Adv. Res.) is an applied/natural sciences, peer-reviewed journal that focuses on interdisciplinary research. The journal aims to contribute to applied research and knowledge worldwide through the publication of original and high-quality research articles in the fields of Medicine, Pharmaceutical Sciences, Dentistry, Physical Therapy, Veterinary Medicine, and Basic and Biological Sciences.
The following abstracting and indexing services cover the Journal of Advanced Research: PubMed/Medline, Essential Science Indicators, Web of Science, Scopus, PubMed Central, PubMed, Science Citation Index Expanded, Directory of Open Access Journals (DOAJ), and INSPEC.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


