A palmitoylation–depalmitoylation relay spatiotemporally controls GSDMD activation in pyroptosis
IF 17.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
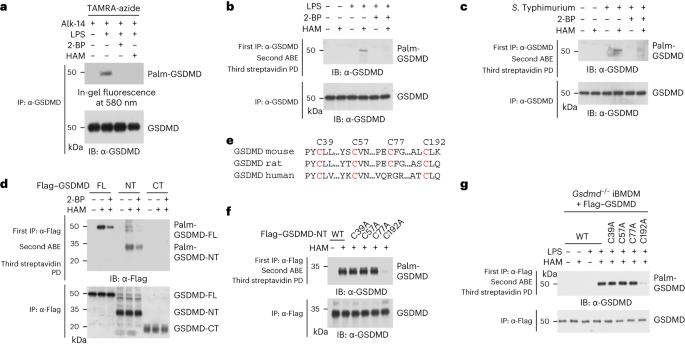
Gasdermin D (GSDMD) is the executor of pyroptosis, which is important for host defence against pathogen infection. Following activation, caspase-mediated cleavage of GSDMD releases an amino-terminal fragment (GSDMD-NT), which oligomerizes and forms pores in the plasma membrane, leading to cell death and release of proinflammatory cytokines. The spatial and temporal regulation of this process in cells remains unclear. Here we identify GSDMD as a substrate for reversible S-palmitoylation on C192 during pyroptosis. The palmitoyl acyltransferase DHHC7 palmitoylates GSDMD to direct its cleavage by caspases. Subsequently, palmitoylation of GSDMD-NT promotes its translocation to the plasma membrane, where APT2 depalmitoylates GSDMD-NT to unmask the C192 residue and promote GSDMD-NT oligomerization. Perturbation of either palmitoylation or depalmitoylation suppresses pyroptosis, leading to increased survival of mice with lipopolysaccharide-induced lethal septic shock and increased sensitivity to bacterial infection. Our findings reveal a model through which a palmitoylation–depalmitoylation relay spatiotemporally controls GSDMD activation during pyroptosis. Xu and colleagues identify a sequential palmitoylation–depalmitoylation mechanism that controls GSDMD cleavage by caspases, plasma membrane trafficking and oligomerization, thereby triggering pyroptosis in a spatial and temporal manner.
棕榈酰化-去棕榈酰化中继在时空上控制着热核病中 GSDMD 的激活
Gasdermin D(GSDMD)是热核变性的执行者,对宿主抵御病原体感染非常重要。激活后,Caspase 介导的 GSDMD 裂解释放出一个氨基末端片段(GSDMD-NT),该片段寡聚并在质膜上形成孔,导致细胞死亡并释放促炎细胞因子。细胞中这一过程的空间和时间调控仍不清楚。在这里,我们确定 GSDMD 是热核分裂过程中 C192 上可逆 S-棕榈酰化的底物。棕榈酰酰基酰基转移酶 DHHC7 对 GSDMD 进行棕榈酰化,以引导 Caspases 对其进行裂解。随后,GSDMD-NT 的棕榈酰化促进其转运至质膜,APT2 在质膜上对 GSDMD-NT 进行去棕榈酰化,以解除 C192 残基的屏蔽并促进 GSDMD-NT 的寡聚化。棕榈酰化或去棕榈酰化的干扰都会抑制热渗透,导致脂多糖诱导的致死性败血性休克小鼠存活率提高,对细菌感染的敏感性增加。我们的研究结果揭示了一个模型,通过该模型,棕榈酰化-去棕榈酰化中继在热蛋白沉积过程中对 GSDMD 的激活进行时空控制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


