Towards a general-purpose foundation model for computational pathology
IF 58.7
1区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
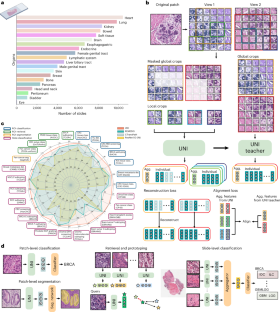
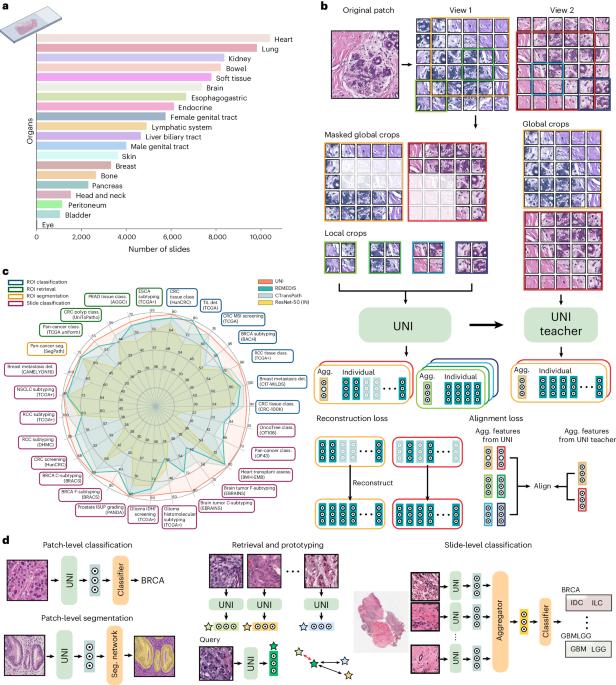
Quantitative evaluation of tissue images is crucial for computational pathology (CPath) tasks, requiring the objective characterization of histopathological entities from whole-slide images (WSIs). The high resolution of WSIs and the variability of morphological features present significant challenges, complicating the large-scale annotation of data for high-performance applications. To address this challenge, current efforts have proposed the use of pretrained image encoders through transfer learning from natural image datasets or self-supervised learning on publicly available histopathology datasets, but have not been extensively developed and evaluated across diverse tissue types at scale. We introduce UNI, a general-purpose self-supervised model for pathology, pretrained using more than 100 million images from over 100,000 diagnostic H&E-stained WSIs (>77 TB of data) across 20 major tissue types. The model was evaluated on 34 representative CPath tasks of varying diagnostic difficulty. In addition to outperforming previous state-of-the-art models, we demonstrate new modeling capabilities in CPath such as resolution-agnostic tissue classification, slide classification using few-shot class prototypes, and disease subtyping generalization in classifying up to 108 cancer types in the OncoTree classification system. UNI advances unsupervised representation learning at scale in CPath in terms of both pretraining data and downstream evaluation, enabling data-efficient artificial intelligence models that can generalize and transfer to a wide range of diagnostically challenging tasks and clinical workflows in anatomic pathology. Pretrained using over 100,000 diagnostic histopathological slides across 20 major tissue types, a self-supervised model is shown to outperform existing baselines across various clinically relevant computational pathology tasks.
为计算病理学建立通用基础模型。
组织图像的定量评估对于计算病理学(CPath)任务至关重要,它要求从全切片图像(WSI)中客观地描述组织病理学实体。WSIs 的高分辨率和形态特征的多变性带来了巨大的挑战,使得为高性能应用进行大规模数据注释变得更加复杂。为了应对这一挑战,目前的研究提出了通过自然图像数据集的迁移学习或公开组织病理学数据集的自监督学习来使用预训练图像编码器,但这些方法尚未在不同组织类型中进行大规模的广泛开发和评估。我们介绍的 UNI 是一种通用的病理自监督模型,它使用来自 20 种主要组织类型的 10 万多张诊断性 H&E 染色 WSI 的 1 亿多幅图像(>77 TB 的数据)进行预训练。该模型在 34 个具有代表性的不同诊断难度的 CPath 任务中进行了评估。除了表现优于以前的一流模型外,我们还展示了 CPath 中的新建模能力,如分辨率失真组织分类、使用少镜头类原型进行切片分类,以及在 OncoTree 分类系统中对多达 108 种癌症类型进行疾病亚型归纳分类。UNI 推进了 CPath 在预训练数据和下游评估方面的大规模无监督表征学习,使数据高效的人工智能模型能够泛化并转移到解剖病理学中各种具有诊断挑战性的任务和临床工作流程中。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Medicine
医学-生化与分子生物学
CiteScore
100.90
自引率
0.70%
发文量
525
审稿时长
1 months
期刊介绍:
Nature Medicine is a monthly journal publishing original peer-reviewed research in all areas of medicine. The publication focuses on originality, timeliness, interdisciplinary interest, and the impact on improving human health. In addition to research articles, Nature Medicine also publishes commissioned content such as News, Reviews, and Perspectives. This content aims to provide context for the latest advances in translational and clinical research, reaching a wide audience of M.D. and Ph.D. readers. All editorial decisions for the journal are made by a team of full-time professional editors.
Nature Medicine consider all types of clinical research, including:
-Case-reports and small case series
-Clinical trials, whether phase 1, 2, 3 or 4
-Observational studies
-Meta-analyses
-Biomarker studies
-Public and global health studies
Nature Medicine is also committed to facilitating communication between translational and clinical researchers. As such, we consider “hybrid” studies with preclinical and translational findings reported alongside data from clinical studies.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


