Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX
IF 16
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
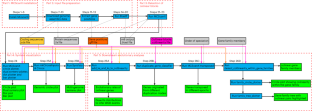
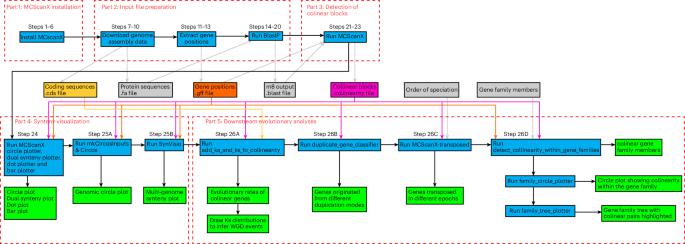
As different taxa evolve, gene order often changes slowly enough that chromosomal ‘blocks’ with conserved gene orders (synteny) are discernible. The MCScanX toolkit ( https://github.com/wyp1125/MCScanX ) was published in 2012 as freely available software for the detection of such ‘colinear blocks’ and subsequent synteny and evolutionary analyses based on genome-wide gene location and protein sequence information. Owing to its simplicity and high efficiency for colinear block detection, MCScanX provides a powerful tool for conducting diverse synteny and evolutionary analyses. Moreover, the detection of colinear blocks has been embraced as an integral step for pangenome graph construction. Here, new application trends of MCScanX are explored, striving to better connect this increasingly used tool to other tools and accelerate insight generation from exponentially growing sequence data. We provide a detailed protocol that covers how to install MCScanX on diverse platforms, tune parameters, prepare input files from data from the National Center for Biotechnology Information, run MCScanX and its visualization and evolutionary analysis tools, and connect MCScanX with external tools, including MCScanX-transposed, Circos and SynVisio. This protocol is easily implemented by users with minimal computational background and is adaptable to new data of interest to them. The data and utility programs for this protocol can be obtained from http://bdx-consulting.com/mcscanx-protocol . Synteny and colinearity are important parameters that delineate the evolution of genomes and gene families. This protocol describes MCScanX, a user-friendly toolkit that facilitates rapid evolutionary analysis of chromosomal structural changes.
利用 MCScanX 检测共线区块和同源关系,并进行进化分析。
在不同类群的进化过程中,基因顺序的变化往往非常缓慢,以至于基因顺序一致(同源)的染色体 "区块 "可以被分辨出来。MCScanX 工具包 ( https://github.com/wyp1125/MCScanX ) 于 2012 年发布,是一款可免费使用的软件,用于检测此类 "共线区块",并根据全基因组的基因位置和蛋白质序列信息进行后续的同源关系和进化分析。由于 MCScanX 在检测共线性区块方面的简便性和高效性,它为开展多种多样的同源关系和进化分析提供了强有力的工具。此外,共线区块检测已被视为构建泛基因组图谱的一个不可或缺的步骤。在此,我们探讨了 MCScanX 的新应用趋势,力图更好地将这一日益常用的工具与其他工具连接起来,并加速从指数级增长的序列数据中产生洞察力。我们提供了一个详细的协议,包括如何在不同平台上安装 MCScanX、调整参数、从美国国家生物技术信息中心的数据中准备输入文件、运行 MCScanX 及其可视化和进化分析工具,以及将 MCScanX 与外部工具(包括 MCScanX-transposed、Circos 和 SynVisio)连接起来。用户只需具备最低限度的计算背景,就能轻松实施该协议,并能适应他们感兴趣的新数据。该协议的数据和实用程序可从 http://bdx-consulting.com/mcscanx-protocol 获取。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


