Motion Corrected DCE-MR Image Reconstruction Using Deep Learning
Abstract
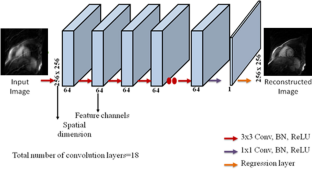
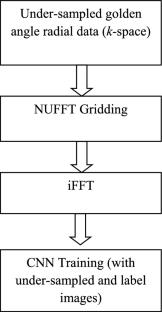
Respiratory motion in abdomen generates motion artifacts during Dynamic Contrast Enhanced MRI (DCE-MRI) data acquisition and it is clinically challenging to minimize the motion artifacts. Extraction of self-gated respiratory signal from the acquired k-space data is one of the methods to deal with the respiratory motion artifacts. The literature shows that non-Cartesian trajectories are less sensitive to motion artifacts than Cartesian trajectories. Golden-angle data acquisition in radial trajectory is preferred to extract the self-gated signal that splits the free-breathing data into different respiratory phases; also called motion states or bins. Conventionally, XD-GRASP (eXtra-Dimension golden-angle-radial Sparse Parallel MRI) reconstructs the binned data, but this method has limitations such as it does not preserve noise like texture (MR images have noise like artifacts) and it is a computationally intensive method. This research work proposes the use of a dedicated Convolutional Neural Network (CNN) architecture to remove motion artifacts from the binned (using uniform and adaptive binning) DCE golden-angle-radial liver perfusion data. The results of the proposed method are compared with XD-GRASP reconstruction. The results demonstrate that the proposed method takes significantly less computation time and provides similar quality of the reconstructed images as compared to the XD-GRASP method. Furthermore, receiver coil sensitivity information is required in XD-GRASP to reconstruct the MR image that may be difficult to estimate in some applications, whereas the proposed method does not require any such information.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


