Regulatory activity is the default DNA state in eukaryotes
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Genomes encode for genes and non-coding DNA, both capable of transcriptional activity. However, unlike canonical genes, many transcripts from non-coding DNA have limited evidence of conservation or function. Here, to determine how much biological noise is expected from non-genic sequences, we quantify the regulatory activity of evolutionarily naive DNA using RNA-seq in yeast and computational predictions in humans. In yeast, more than 99% of naive DNA bases were transcribed. Unlike the evolved transcriptome, naive transcripts frequently overlapped with opposite sense transcripts, suggesting selection favored coherent gene structures in the yeast genome. In humans, regulation-associated chromatin activity is predicted to be common in naive dinucleotide-content-matched randomized DNA. Here, naive and evolved DNA have similar co-occurrence and cell-type specificity of chromatin marks, challenging these as indicators of selection. However, in both yeast and humans, extreme high activities were rare in naive DNA, suggesting they result from selection. Overall, basal regulatory activity seems to be the default, which selection can hone to evolve a function or, if detrimental, repress. Here, the authors ask how much regulatory activity DNA is expected to have in the absence of selection. In yeast and humans, they find that gene regulatory activity is common in evolutionarily naive DNA, suggesting that activity is not always indicative of function.
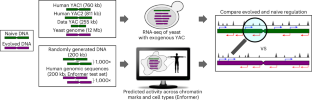
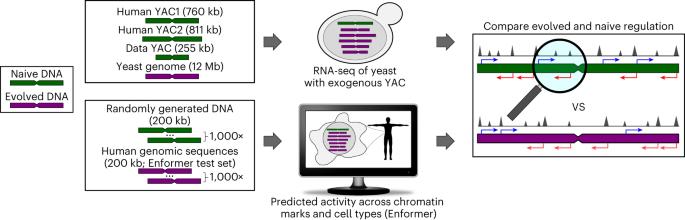
调控活性是真核生物 DNA 的默认状态
基因组编码基因和非编码 DNA,两者都能进行转录活动。然而,与典型基因不同,许多来自非编码 DNA 的转录本的保护或功能证据有限。为了确定非基因序列会产生多少生物噪音,我们利用酵母中的 RNA-seq 和人类中的计算预测对进化中的非编码 DNA 的调控活性进行了量化。在酵母中,99% 以上的幼稚 DNA 碱基被转录。与进化转录组不同的是,幼稚转录本经常与相反的意义转录本重叠,这表明酵母基因组中的选择有利于连贯的基因结构。在人类中,与调控相关的染色质活性被预测为常见于天真二核苷酸内容匹配的随机DNA中。在这里,幼稚DNA和进化DNA的染色质标记具有相似的共存性和细胞类型特异性,这对作为选择指标的染色质标记提出了挑战。然而,在酵母和人类中,极高的活性在幼稚DNA中都很罕见,这表明它们是选择的结果。总之,基础调控活性似乎是默认的,选择可以磨练这种活性,使其进化出某种功能,如果有害,则加以抑制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


