Potential drug targets for gastroesophageal reflux disease and Barrett’s esophagus identified through Mendelian randomization analysis
IF 2.6
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
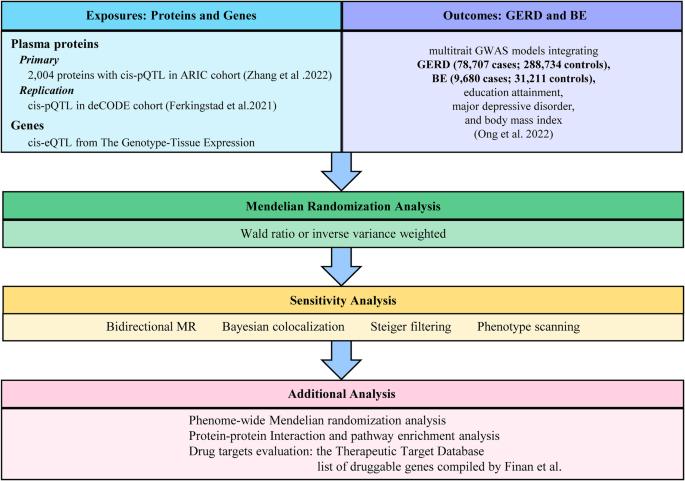
Gastroesophageal reflux disease (GERD) is a prevalent chronic ailment, and present therapeutic approaches are not always effective. This study aimed to find new drug targets for GERD and Barrett’s esophagus (BE). We obtained genetic instruments for GERD, BE, and 2004 plasma proteins from recently published genome-wide association studies (GWAS), and Mendelian randomization (MR) was employed to explore potential drug targets. We further winnowed down MR-prioritized proteins through replication, reverse causality testing, colocalization analysis, phenotype scanning, and Phenome-wide MR. Furthermore, we constructed a protein-protein interaction network, unveiling potential associations among candidate proteins. Simultaneously, we acquired mRNA expression quantitative trait loci (eQTL) data from another GWAS encompassing four different tissues to identify additional drug targets. Meanwhile, we searched drug databases to evaluate these targets. Under Bonferroni correction (P < 4.8 × 10−5), we identified 11 plasma proteins significantly associated with GERD. Among these, 7 are protective proteins (MSP, GPX1, ERBB3, BT3A3, ANTR2, CCM2, and DECR2), while 4 are detrimental proteins (TMEM106B, DUSP13, C1-INH, and LINGO1). Ultimately, C1-INH and DECR2 successfully passed the screening process and exhibited similar directional causal effects on BE. Further analysis of eQTLs highlighted 4 potential drug targets, including EDEM3, PBX3, MEIS1-AS3, and NME7. The search of drug databases further supported our conclusions. Our study indicated that the plasma proteins C1-INH and DECR2, along with 4 genes (EDEM3, PBX3, MEIS1-AS3, and NME7), may represent potential drug targets for GERD and BE, warranting further investigation.
通过孟德尔随机分析确定胃食管反流病和巴雷特食管的潜在药物靶点
胃食管反流病(GERD)是一种普遍存在的慢性疾病,目前的治疗方法并不总是有效。本研究旨在寻找治疗胃食管反流病和巴雷特食管(BE)的新药靶点。我们从最近发表的全基因组关联研究(GWAS)中获得了胃食管反流病、巴雷特食管炎和 2004 年血浆蛋白的遗传工具,并采用孟德尔随机化法(MR)探索潜在的药物靶点。我们通过复制、反向因果关系测试、共聚焦分析、表型扫描和全表型 MR 进一步筛选出 MR 优先选择的蛋白质。此外,我们还构建了蛋白质-蛋白质相互作用网络,揭示了候选蛋白质之间的潜在关联。与此同时,我们还从另一个涵盖四个不同组织的 GWAS 中获取了 mRNA 表达定量性状位点(eQTL)数据,以确定更多的药物靶点。同时,我们还搜索了药物数据库以评估这些靶点。在 Bonferroni 校正(P < 4.8 × 10-5)下,我们确定了 11 种与胃食管反流病显著相关的血浆蛋白。其中,7 种是保护性蛋白(MSP、GPX1、ERBB3、BT3A3、ANTR2、CCM2 和 DECR2),4 种是有害蛋白(TMEM106B、DUSP13、C1-INH 和 LINGO1)。最终,C1-INH 和 DECR2 顺利通过筛选,并对 BE 表现出相似的定向因果效应。对eQTLs的进一步分析突出了4个潜在的药物靶点,包括EDEM3、PBX3、MEIS1-AS3和NME7。对药物数据库的搜索进一步支持了我们的结论。我们的研究表明,血浆蛋白 C1-INH 和 DECR2 以及 4 个基因(EDEM3、PBX3、MEIS1-AS3 和 NME7)可能是治疗胃食管反流病和 BE 的潜在药物靶点,值得进一步研究。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


