Deadenylation kinetics of mixed poly(A) tails at single-nucleotide resolution
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
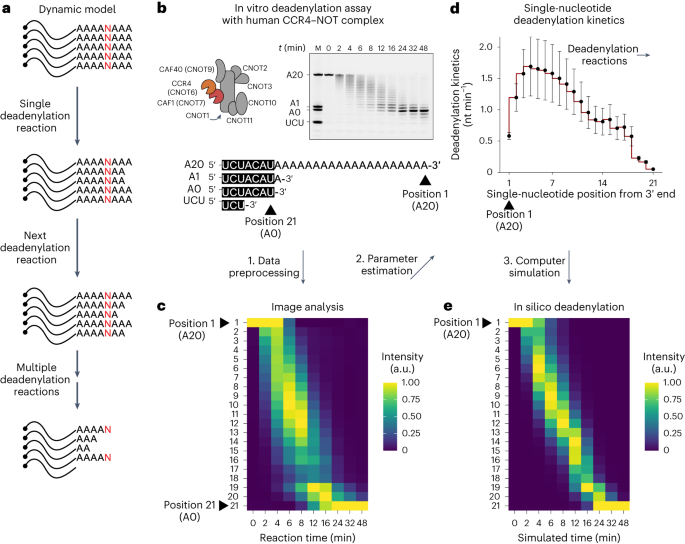
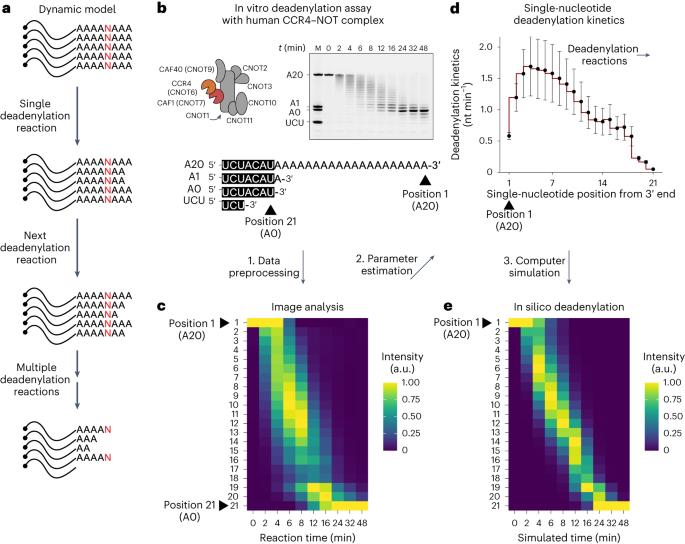
Shortening of messenger RNA poly(A) tails, or deadenylation, is a rate-limiting step in mRNA decay and is highly regulated during gene expression. The incorporation of non-adenosines in poly(A) tails, or ‘mixed tailing’, has been observed in vertebrates and viruses. Here, to quantitate the effect of mixed tails, we mathematically modeled deadenylation reactions at single-nucleotide resolution using an in vitro deadenylation system reconstituted with the complete human CCR4–NOT complex. Applying this model, we assessed the disrupting impact of single guanosine, uridine or cytosine to be equivalent to approximately 6, 8 or 11 adenosines, respectively. CCR4–NOT stalls at the 0, −1 and −2 positions relative to the non-adenosine residue. CAF1 and CCR4 enzyme subunits commonly prefer adenosine but exhibit distinct sequence selectivities and stalling positions. Our study provides an analytical framework to monitor deadenylation and reveals the molecular basis of tail sequence-dependent regulation of mRNA stability. Here, using mathematical modeling and an in vitro deadenylation system, the authors quantitatively demonstrate the effect of non-adenosines in the poly(A) tail and exemplify how tail sequence regulates mRNA stability.
单核苷酸分辨率下混合聚(A)尾的去烯化动力学
信使 RNA 聚(A)尾的缩短或去腺苷化是 mRNA 衰减的限速步骤,在基因表达过程中受到高度调控。在脊椎动物和病毒中已经观察到非腺苷在聚(A)尾中的结合,即 "混合尾化"。在这里,为了量化混合尾的影响,我们使用一个由完整的人类 CCR4-NOT 复合物重组的体外去腺苷化系统,以单核苷酸分辨率对去腺苷化反应进行了数学建模。应用该模型,我们评估了单个鸟苷、尿苷或胞嘧啶的破坏性影响,它们分别相当于约 6、8 或 11 个腺苷。相对于非腺苷残基,CCR4-NOT 在 0、-1 和 -2 位停滞。CAF1 和 CCR4 酶亚基通常偏好腺苷,但表现出不同的序列选择性和停滞位置。我们的研究提供了一个监测去腺苷化的分析框架,并揭示了尾序列依赖性调控 mRNA 稳定性的分子基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
文献相关原料
| 公司名称 | 产品信息 | 采购帮参考价格 |
|---|
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


