Cis-regulatory control of mammalian Trps1 gene expression
Abstract
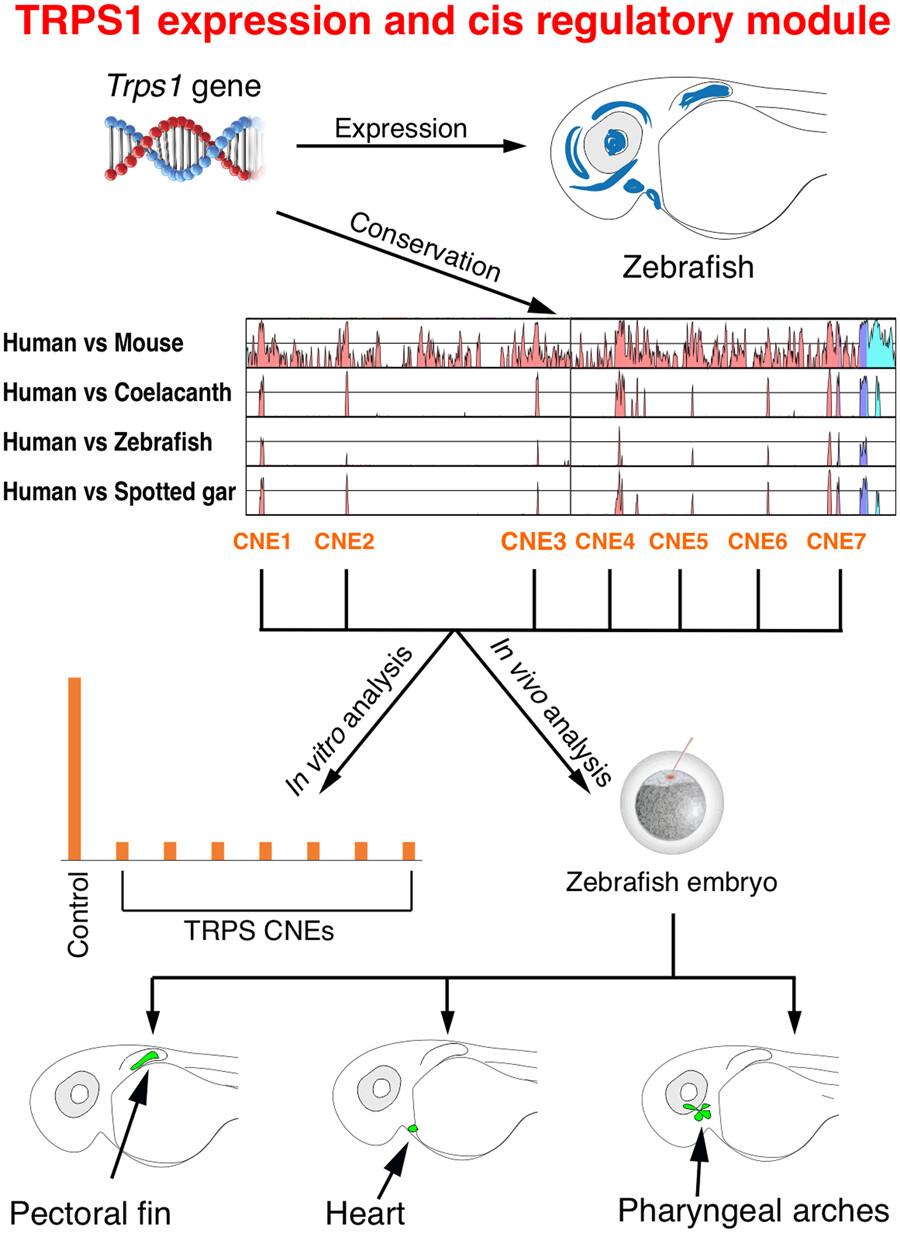
TRPS1 serves as the causative gene for tricho-rhino phalangeal syndrome, known for its craniofacial and skeletal abnormalities. The Trps1 gene encodes a protein that represses Wnt signaling through strong interactions with Wnt signaling inhibitors. The identification of genomic cis-acting regulatory sequences governing Trps1 expression is crucial for understanding its role in embryogenesis. Nevertheless, to date, no investigations have been conducted concerning these aspects of Trps1. To identify deeply conserved noncoding elements (CNEs) within the Trps1 locus, we employed a comparative genomics approach, utilizing slowly evolving fish such as coelacanth and spotted gar. These analyses resulted in the identification of eight CNEs in the intronic region of the Trps1 gene. Functional characterization of these CNEs in zebrafish revealed their regulatory potential in various tissues, including pectoral fins, heart, and pharyngeal arches. RNA in-situ hybridization experiments revealed concordance between the reporter expression pattern induced by the identified set of CNEs and the spatial expression pattern of the trps1 gene in zebrafish. Comparative in vivo data from zebrafish and mice for CNE7/hs919 revealed conserved functions of these enhancers. Each of these eight CNEs was further investigated in cell line-based reporter assays, revealing their repressive potential. Taken together, in vivo and in vitro assays suggest a context-dependent dual functionality for the identified set of Trps1-associated CNE enhancers. This functionally characterized set of CNE-enhancers will contribute to a more comprehensive understanding of the developmental roles of Trps1 and can aid in the identification of noncoding DNA variants associated with human diseases.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


