A non-invasive 25-Gene PLNM-Score urine test for detection of prostate cancer pelvic lymph node metastasis
IF 5.1
2区 医学
Q1 ONCOLOGY
引用次数: 0
Abstract
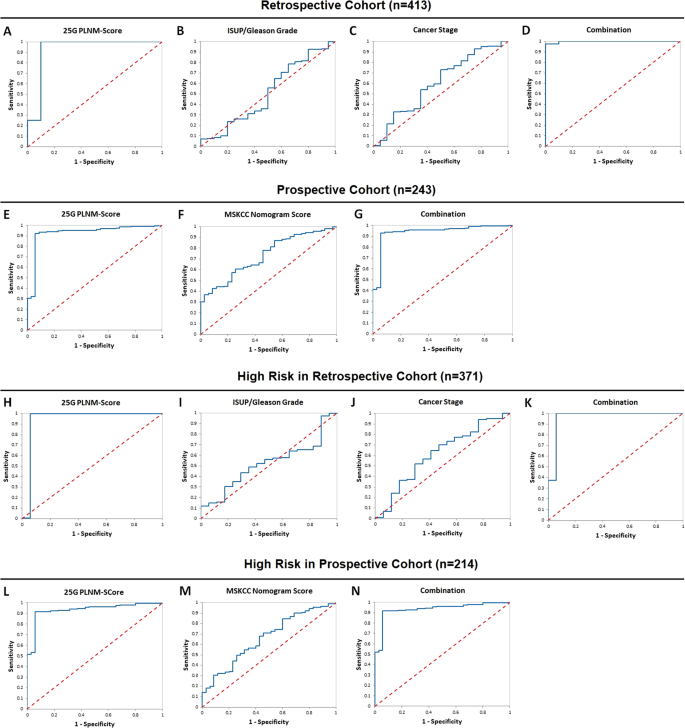
Prostate cancer patients with pelvic lymph node metastasis (PLNM) have poor prognosis. Based on EAU guidelines, patients with >5% risk of PLNM by nomograms often receive pelvic lymph node dissection (PLND) during prostatectomy. However, nomograms have limited accuracy, so large numbers of false positive patients receive unnecessary surgery with potentially serious side effects. It is important to accurately identify PLNM, yet current tests, including imaging tools are inaccurate. Therefore, we intended to develop a gene expression-based algorithm for detecting PLNM. An advanced random forest machine learning algorithm screening was conducted to develop a classifier for identifying PLNM using urine samples collected from a multi-center retrospective cohort (n = 413) as training set and validated in an independent multi-center prospective cohort (n = 243). Univariate and multivariate discriminant analyses were performed to measure the ability of the algorithm classifier to detect PLNM and compare it with the Memorial Sloan Kettering Cancer Center (MSKCC) nomogram score. An algorithm named 25 G PLNM-Score was developed and found to accurately distinguish PLNM and non-PLNM with AUC of 0.93 (95% CI: 0.85–1.01) and 0.93 (95% CI: 0.87–0.99) in the retrospective and prospective urine cohorts respectively. Kaplan–Meier plots showed large and significant difference in biochemical recurrence-free survival and distant metastasis-free survival in the patients stratified by the 25 G PLNM-Score (log rank P < 0.001 and P < 0.0001, respectively). It spared 96% and 80% of unnecessary PLND with only 0.51% and 1% of PLNM missing in the retrospective and prospective cohorts respectively. In contrast, the MSKCC score only spared 15% of PLND with 0% of PLNM missing. The novel 25 G PLNM-Score is the first highly accurate and non-invasive machine learning algorithm-based urine test to identify PLNM before PLND, with potential clinical benefits of avoiding unnecessary PLND and improving treatment decision-making.

用于检测前列腺癌盆腔淋巴结转移的无创 25 基因 PLNM-Score 尿液检验
背景有盆腔淋巴结转移(PLNM)的前列腺癌患者预后较差。根据欧洲前列腺癌联盟(EAU)的指南,根据提名图显示有5% PLNM风险的患者通常会在前列腺切除术中接受盆腔淋巴结清扫术(PLND)。然而,提名图的准确性有限,因此大量假阳性患者接受了不必要的手术,并可能带来严重的副作用。准确识别前列腺淋巴结核非常重要,但目前的检测方法(包括成像工具)并不准确。方法使用从多中心回顾性队列(n = 413)中收集的尿液样本作为训练集,并在独立的多中心前瞻性队列(n = 243)中进行验证,采用先进的随机森林机器学习算法筛选,开发出识别 PLNM 的分类器。结果 开发出一种名为25 G PLNM-Score的算法,该算法能准确区分PLNM和非PLNM,在回顾性和前瞻性尿液队列中的AUC分别为0.93(95% CI:0.85-1.01)和0.93(95% CI:0.87-0.99)。Kaplan-Meier图显示,按25 G PLNM-Score分层的患者无生化复发生存率和无远处转移生存率差异巨大且显著(对数秩分别为P < 0.001和P < 0.0001)。在回顾性队列和前瞻性队列中,分别只有0.51%和1%的PLNM缺失,避免了96%和80%不必要的PLND。结论新颖的25 G PLNM-Score是首个基于机器学习算法的高精度、无创尿液检验,可在PLND前识别PLNM,具有避免不必要的PLND和改善治疗决策的潜在临床益处。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Prostate Cancer and Prostatic Diseases
医学-泌尿学与肾脏学
CiteScore
10.00
自引率
6.20%
发文量
142
审稿时长
6-12 weeks
期刊介绍:
Prostate Cancer and Prostatic Diseases covers all aspects of prostatic diseases, in particular prostate cancer, the subject of intensive basic and clinical research world-wide. The journal also reports on exciting new developments being made in diagnosis, surgery, radiotherapy, drug discovery and medical management.
Prostate Cancer and Prostatic Diseases is of interest to surgeons, oncologists and clinicians treating patients and to those involved in research into diseases of the prostate. The journal covers the three main areas - prostate cancer, male LUTS and prostatitis.
Prostate Cancer and Prostatic Diseases publishes original research articles, reviews, topical comment and critical appraisals of scientific meetings and the latest books. The journal also contains a calendar of forthcoming scientific meetings. The Editors and a distinguished Editorial Board ensure that submitted articles receive fast and efficient attention and are refereed to the highest possible scientific standard. A fast track system is available for topical articles of particular significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


