Methane-dependent complete denitrification by a single Methylomirabilis bacterium
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
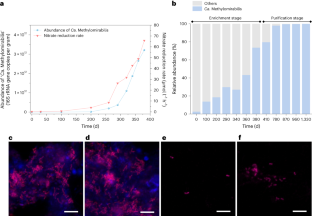
Methane-dependent nitrate and nitrite removal in anoxic environments is thought to rely on syntrophy between ANME-2d archaea and bacteria in the genus ‘Candidatus Methylomirabilis’. Here we enriched and purified a single Methylomirabilis from paddy soil fed with nitrate and methane, which is capable of coupling methane oxidation to nitrate reduction via nitrite to dinitrogen independently. Isotope labelling showed that this bacterium we name ‘Ca. Methylomirabilis sinica’ stoichiometrically performed methane-dependent complete nitrate reduction to dinitrogen gas. Multi-omics analyses collectively demonstrated that ‘M. sinica’ actively expressed a well-established pathway for this process, especially including nitrate reductase Nap. Furthermore, ‘M. sinica’ exhibited a higher nitrate affinity than most denitrifiers, implying its competitive fitness under oligotrophic nitrogen-limited conditions. Our findings revise the paradigm of methane-dependent denitrification performed by two organisms, and the widespread presence of ‘M. sinica’ in public databases suggests that the coupling of methane oxidation and complete denitrification in single cells substantially contributes to global methane and nitrogen budgets. Enrichments from bioreactors find a single bacterium capable of methane oxidation coupled to nitrate reduction via nitrite to dinitrogen, a process previously thought to rely on syntrophy.
单个 Methylomirabilis 细菌的甲烷依赖性完全反硝化作用
缺氧环境中甲烷依赖性硝酸盐和亚硝酸盐的去除被认为依赖于 ANME-2d 古细菌和 "Candidatus Methylomirabilis "属细菌之间的合成作用。在这里,我们从含有硝酸盐和甲烷的稻田土壤中富集并纯化了一种单一的 Methylomirabilis,它能够独立地将甲烷氧化与硝酸盐还原(通过亚硝酸盐还原成二氮)耦合在一起。同位素标记显示,这种细菌被我们命名为 "Ca.Methylomirabilis sinica "的细菌在甲烷依赖性硝酸盐完全还原为二氮气的过程中达到了相同的化学计量。多组学分析共同表明,'M. sinica'积极表达了这一过程的成熟途径,特别是包括硝酸还原酶Nap。此外,'M. sinica'比大多数反硝化细菌表现出更高的硝酸盐亲和力,这意味着它在寡营养氮限制条件下具有竞争优势。我们的发现修正了由两种生物进行甲烷依赖性反硝化作用的范式,'M. sinica'在公共数据库中的广泛存在表明,单细胞中甲烷氧化和完全反硝化作用的耦合对全球甲烷和氮预算做出了重大贡献。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


