Detection of hidden intronic DDC variant in aromatic L-amino acid decarboxylase deficiency by adaptive sampling
IF 2.6
3区 生物学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
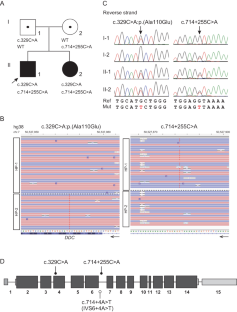
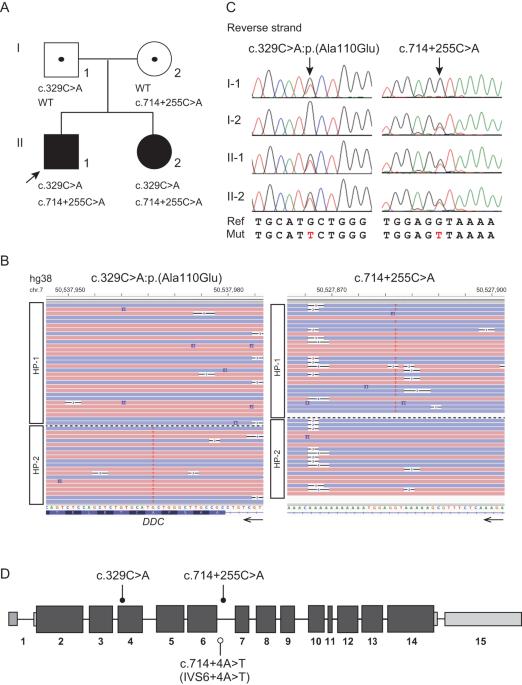
Aromatic l-amino acid decarboxylase (AADC) deficiency is an autosomal recessive neurotransmitter disorder caused by pathogenic DOPA decarboxylase (DDC) variants. We previously reported Japanese siblings with AADC deficiency, which was confirmed by the lack of enzyme activity; however, only a heterozygous missense variant was detected. We therefore performed targeted long-read sequencing by adaptive sampling to identify any missing variants. Haplotype phasing and variant calling identified a novel deep intronic variant (c.714+255 C > A), which was predicted to potentially activate the noncanonical splicing acceptor site. Minigene assay revealed that wild-type and c.714+255 C > A alleles had different impacts on splicing. Three transcripts, including the canonical transcript, were detected from the wild-type allele, but only the noncanonical cryptic exon was produced from the variant allele, indicating that c.714+255 C > A was pathogenic. Target long-read sequencing may be used to detect hidden pathogenic variants in unresolved autosomal recessive cases with only one disclosed hit variant.
通过适应性取样检测芳香族 L-氨基酸脱羧酶缺乏症中隐藏的内含子 DDC 变异。
芳香族l-氨基酸脱羧酶(AADC)缺乏症是一种常染色体隐性神经递质紊乱,由致病性DOPA脱羧酶(DDC)变体引起。我们以前曾报道过患有 AADC 缺乏症的日本兄妹,他们因缺乏酶活性而被确诊;然而,只检测到了一个杂合子错义变体。因此,我们通过自适应采样进行了有针对性的长线程测序,以确定任何缺失的变异。单倍型分期和变异调用确定了一个新的深内含子变异(c.714+255 C > A),据预测,该变异可能会激活非规范剪接接受位点。迷你基因测定显示,野生型和 c.714+255 C > A 等位基因对剪接的影响不同。从野生型等位基因中检测到三个转录本,包括规范转录本,但从变异等位基因中只产生了非规范隐性外显子,这表明 c.714+255 C > A 是致病的。靶向长读测序可用于检测仅有一个已知变异的常染色体隐性病例中隐藏的致病变异。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Human Genetics
生物-遗传学
CiteScore
7.20
自引率
0.00%
发文量
101
审稿时长
4-8 weeks
期刊介绍:
The Journal of Human Genetics is an international journal publishing articles on human genetics, including medical genetics and human genome analysis. It covers all aspects of human genetics, including molecular genetics, clinical genetics, behavioral genetics, immunogenetics, pharmacogenomics, population genetics, functional genomics, epigenetics, genetic counseling and gene therapy.
Articles on the following areas are especially welcome: genetic factors of monogenic and complex disorders, genome-wide association studies, genetic epidemiology, cancer genetics, personal genomics, genotype-phenotype relationships and genome diversity.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


