Exploring the next generation of antibody–drug conjugates
IF 82.2
1区 医学
Q1 ONCOLOGY
引用次数: 0
Abstract
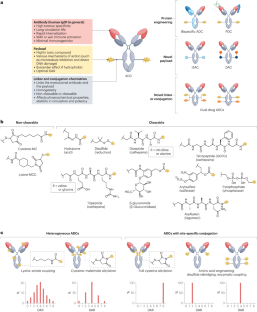
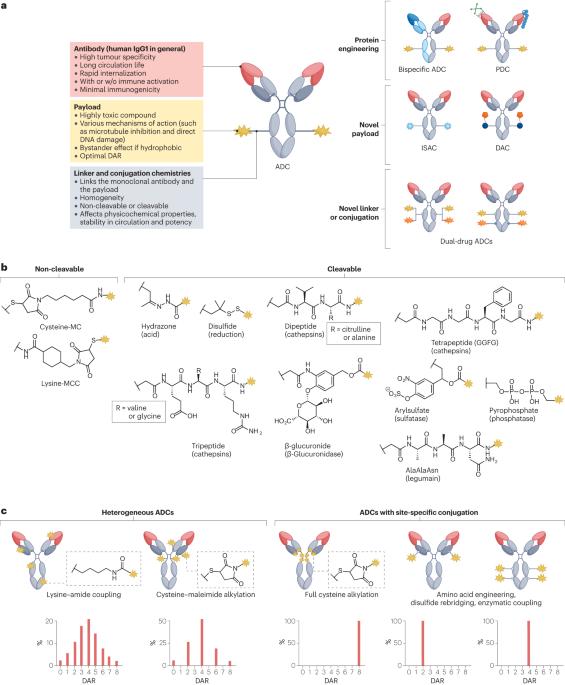
Antibody–drug conjugates (ADCs) are a promising cancer treatment modality that enables the selective delivery of highly cytotoxic payloads to tumours. However, realizing the full potential of this platform necessitates innovative molecular designs to tackle several clinical challenges such as drug resistance, tumour heterogeneity and treatment-related adverse effects. Several emerging ADC formats exist, including bispecific ADCs, conditionally active ADCs (also known as probody–drug conjugates), immune-stimulating ADCs, protein-degrader ADCs and dual-drug ADCs, and each offers unique capabilities for tackling these various challenges. For example, probody–drug conjugates can enhance tumour specificity, whereas bispecific ADCs and dual-drug ADCs can address resistance and heterogeneity with enhanced activity. The incorporation of immune-stimulating and protein-degrader ADCs, which have distinct mechanisms of action, into existing treatment strategies could enable multimodal cancer treatment. Despite the promising outlook, the importance of patient stratification and biomarker identification cannot be overstated for these emerging ADCs, as these factors are crucial to identify patients who are most likely to derive benefit. As we continue to deepen our understanding of tumour biology and refine ADC design, we will edge closer to developing truly effective and safe ADCs for patients with treatment-refractory cancers. In this Review, we highlight advances in each ADC component (the monoclonal antibody, payload, linker and conjugation chemistry) and provide more-detailed discussions on selected examples of emerging novel ADCs of each format, enabled by engineering of one or more of these components. Antibody–drug conjugates (ADCs) are effective cancer drugs that have been approved for more than 20 specific indications. Nonetheless, acquired resistance and adverse events both limit the effectiveness of these agents. In this Review, the authors describe the development of novel ADC designs, including bispecific ADCs, probody–drug conjugates, immune-stimulating ADCs, protein-degrader ADCs and dual-drug ADCs. all of which have the potential to address these challenges and provide more effective ADCs.
探索新一代抗体药物共轭物
抗体-药物共轭物(ADCs)是一种前景广阔的癌症治疗模式,它能选择性地向肿瘤输送高细胞毒性有效载荷。然而,要充分发挥这一平台的潜力,就必须进行创新的分子设计,以应对耐药性、肿瘤异质性和治疗相关不良反应等若干临床挑战。目前有几种新兴的 ADC 形式,包括双特异性 ADC、条件活性 ADC(也称为原体-药物共轭物)、免疫刺激 ADC、蛋白降解 ADC 和双药 ADC,每种形式都有独特的能力来应对这些挑战。例如,原体-药物共轭物可以增强肿瘤特异性,而双特异性 ADC 和双药 ADC 则可以通过增强活性来解决耐药性和异质性问题。将具有不同作用机制的免疫刺激和蛋白降解 ADC 纳入现有的治疗策略,可以实现多模式癌症治疗。尽管前景看好,但患者分层和生物标志物鉴定对于这些新兴 ADCs 的重要性怎么强调都不为过,因为这些因素对于确定最有可能获益的患者至关重要。随着我们不断加深对肿瘤生物学的理解并完善 ADC 设计,我们将更接近为难治性癌症患者开发出真正有效、安全的 ADC。在本综述中,我们将重点介绍 ADC 各个组成部分(单克隆抗体、有效载荷、连接体和共轭化学)的进展,并通过对其中一个或多个组成部分进行工程设计,对每种形式的新兴新型 ADC 示例进行更详细的讨论。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
99.40
自引率
0.40%
发文量
114
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews publishes clinical content authored by internationally renowned clinical academics and researchers, catering to readers in the medical sciences at postgraduate levels and beyond. Although targeted at practicing doctors, researchers, and academics within specific specialties, the aim is to ensure accessibility for readers across various medical disciplines. The journal features in-depth Reviews offering authoritative and current information, contextualizing topics within the history and development of a field. Perspectives, News & Views articles, and the Research Highlights section provide topical discussions, opinions, and filtered primary research from diverse medical journals.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


