BilR is a gut microbial enzyme that reduces bilirubin to urobilinogen
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 2
Abstract
Metabolism of haem by-products such as bilirubin by humans and their gut microbiota is essential to human health, as excess serum bilirubin can cause jaundice and even neurological damage. The bacterial enzymes that reduce bilirubin to urobilinogen, a key step in this pathway, have remained unidentified. Here we used biochemical analyses and comparative genomics to identify BilR as a gut-microbiota-derived bilirubin reductase that reduces bilirubin to urobilinogen. We delineated the BilR sequences from similar reductases through the identification of key residues critical for bilirubin reduction and found that BilR is predominantly encoded by Firmicutes species. Analysis of human gut metagenomes revealed that BilR is nearly ubiquitous in healthy adults, but prevalence is decreased in neonates and individuals with inflammatory bowel disease. This discovery sheds light on the role of the gut microbiome in bilirubin metabolism and highlights the significance of the gut–liver axis in maintaining bilirubin homeostasis. BilR is identified as a bilirubin reductase encoded by gut bacteria, which reduces bilirubin to urobilinogen.
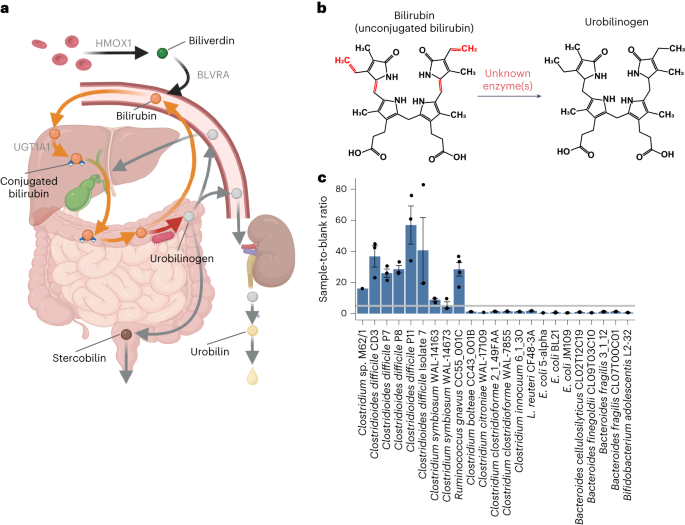
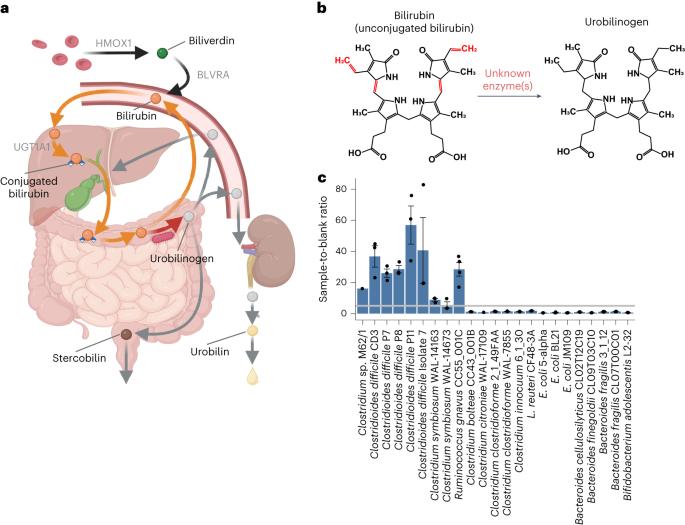
BilR 是一种肠道微生物酶,可将胆红素还原为尿蛋白原
人类及其肠道微生物群对胆红素等血液副产物的代谢对人类健康至关重要,因为血清胆红素过高会导致黄疸,甚至神经损伤。将胆红素还原成尿蛋白原的细菌酶是这一途径的关键步骤,但至今仍未找到。在这里,我们利用生化分析和比较基因组学鉴定出 BilR 是一种来源于肠道微生物群的胆红素还原酶,它能将胆红素还原成尿比林原。我们通过鉴定胆红素还原的关键残基,将 BilR 序列与类似的还原酶区分开来,并发现 BilR 主要由真菌类编码。对人类肠道元基因组的分析表明,BilR 在健康成年人中几乎无处不在,但在新生儿和炎症性肠病患者中的流行率却有所下降。这一发现揭示了肠道微生物组在胆红素代谢中的作用,并强调了肠道-肝脏轴在维持胆红素平衡中的重要性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


