Energy-driven genome regulation by ATP-dependent chromatin remodellers
IF 81.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
The packaging of DNA into chromatin in eukaryotes regulates gene transcription, DNA replication and DNA repair. ATP-dependent chromatin remodelling enzymes (re)arrange nucleosomes at the first level of chromatin organization. Their Snf2-type motor ATPases alter histone–DNA interactions through a common DNA translocation mechanism. Whether remodeller activities mainly catalyse nucleosome dynamics or accurately co-determine nucleosome organization remained unclear. In this Review, we discuss the emerging mechanisms of chromatin remodelling: dynamic remodeller architectures and their interactions, the inner workings of the ATPase cycle, allosteric regulation and pathological dysregulation. Recent mechanistic insights argue for a decisive role of remodellers in the energy-driven self-organization of chromatin, which enables both stability and plasticity of genome regulation — for example, during development and stress. Different remodellers, such as members of the SWI/SNF, ISWI, CHD and INO80 families, process (epi)genetic information through specific mechanisms into distinct functional outputs. Combinatorial assembly of remodellers and their interplay with histone modifications, histone variants, DNA sequence or DNA-bound transcription factors regulate nucleosome mobilization or eviction or histone exchange. Such input–output relationships determine specific nucleosome positions and compositions with distinct DNA accessibilities and mediate differential genome regulation. Finally, remodeller genes are often mutated in diseases characterized by genome dysregulation, notably in cancer, and we discuss their physiological relevance. ATP-dependent chromatin remodellers regulate chromatin transactions — transcription, replication and DNA repair — by re-arranging nucleosomes. Recent studies have elucidated the organization of remodeller complexes, their ATPase activity, their regulation and their pathological dysregulation.
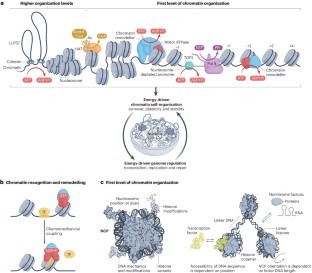
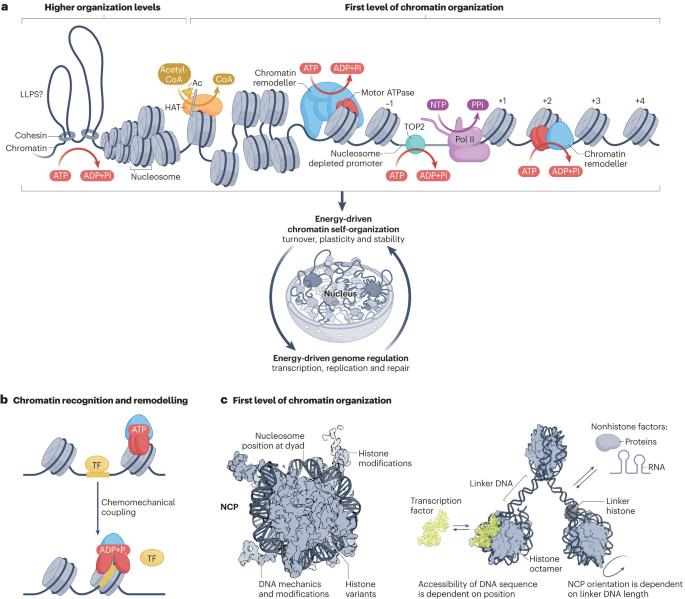
ATP 依赖性染色质重塑器的能量驱动基因组调控
在真核生物中,将 DNA 包装成染色质可调节基因转录、DNA 复制和 DNA 修复。依赖 ATP 的染色质重塑酶在染色质组织的第一层(重新)排列核小体。它们的 Snf2 型马达 ATP 酶通过一种常见的 DNA 易位机制改变组蛋白与 DNA 之间的相互作用。重塑酶的活动是主要催化核小体动力学,还是准确地共同决定核小体的组织,目前仍不清楚。在本综述中,我们将讨论染色质重塑的新机制:动态重塑器结构及其相互作用、ATPase 循环的内部运作、异位调节和病理失调。最新的机理研究表明,染色质重塑器在能量驱动的染色质自组织过程中起着决定性作用,这使得基因组调控的稳定性和可塑性得以实现--例如在发育和应激过程中。不同的重塑因子,如 SWI/SNF、ISWI、CHD 和 INO80 家族成员,通过特定机制将(外)遗传信息加工成不同的功能输出。重组因子的组合装配及其与组蛋白修饰、组蛋白变体、DNA 序列或 DNA 结合的转录因子之间的相互作用,调节着核小体的移动或驱逐或组蛋白交换。这种输入输出关系决定了特定的核小体位置和组成,具有不同的 DNA 可及性,并介导不同的基因组调控。最后,重塑基因常常在以基因组调控失调为特征的疾病(尤其是癌症)中发生突变,我们将讨论它们的生理相关性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
173.60
自引率
0.50%
发文量
118
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Molecular Cell Biology is a prestigious journal that aims to be the primary source of reviews and commentaries for the scientific communities it serves. The journal strives to publish articles that are authoritative, accessible, and enriched with easily understandable figures, tables, and other display items. The goal is to provide an unparalleled service to authors, referees, and readers, and the journal works diligently to maximize the usefulness and impact of each article. Nature Reviews Molecular Cell Biology publishes a variety of article types, including Reviews, Perspectives, Comments, and Research Highlights, all of which are relevant to molecular and cell biologists. The journal's broad scope ensures that the articles it publishes reach the widest possible audience.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


