A target enrichment probe set for resolving phylogenetic relationships in the coffee family, Rubiaceae
Abstract
Premise
Rubiaceae is among the most species-rich plant families, as well as one of the most morphologically and geographically diverse. Currently available phylogenies have mostly relied on few genomic and plastid loci, as opposed to large-scale genomic data. Target enrichment provides the ability to generate sequence data for hundreds to thousands of phylogenetically informative, single-copy loci, which often leads to improved phylogenetic resolution at both shallow and deep taxonomic scales; however, a publicly accessible Rubiaceae-specific probe set that allows for comparable phylogenetic inference across clades is lacking.
Methods
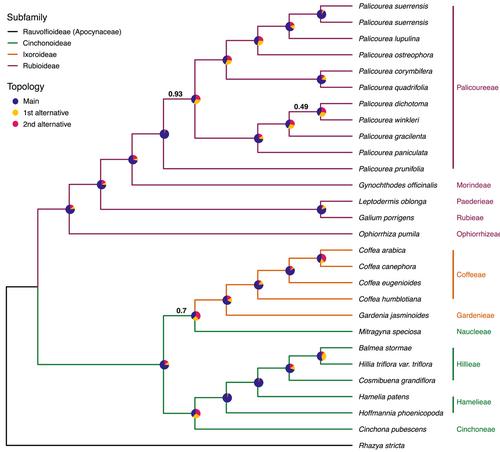
Here, we use publicly accessible genomic resources to identify putatively single-copy nuclear loci for target enrichment in two Rubiaceae groups: tribe Hillieae (Cinchonoideae) and tribal complex Palicoureeae+Psychotrieae (Rubioideae). We sequenced 2270 exonic regions corresponding to 1059 loci in our target clades and generated in silico target enrichment sequences for other Rubiaceae taxa using our designed probe set. To test the utility of our probe set for phylogenetic inference across Rubiaceae, we performed a coalescent-aware phylogenetic analysis using a subset of 27 Rubiaceae taxa from 10 different tribes and three subfamilies, and one outgroup in Apocynaceae.
Results
We recovered an average of 75% and 84% of targeted exons and loci, respectively, per Rubiaceae sample. Probes designed using genomic resources from a particular subfamily were most efficient at targeting sequences from taxa in that subfamily. The number of paralogs recovered during assembly varied for each clade. Phylogenetic inference of Rubiaceae with our target regions resolves relationships at various scales. Relationships are largely consistent with previous studies of relationships in the family with high support (≥0.98 local posterior probability) at nearly all nodes and evidence of gene tree discordance.
Discussion
Our probe set, which we call Rubiaceae2270x, was effective for targeting loci in species across and even outside of Rubiaceae. This probe set will facilitate phylogenomic studies in Rubiaceae and advance systematics and macroevolutionary studies in the family.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


