In silico analysis of a novel hypothetical protein (YP_498675.1) from Staphylococcus aureus unravels the protein of tryptophan synthase beta superfamily (Try-synth-beta_ II)
IF 2.8
Q3 Biochemistry, Genetics and Molecular Biology
Journal of Genetic Engineering and Biotechnology
Pub Date : 2023-12-01
DOI:10.1186/s43141-023-00613-7
引用次数: 0
Abstract
Background
Staphylococcus aureus is a gram-positive spherical bacteria and the most common cause of nosocomial infections in the world. Given its clinical significance, the genome sequence of S. aureus has been elucidated to enhance our comprehension of its lifestyle and pathogenicity. The research aimed to summarize a potential hypothetical protein that may play an important role in S. aureus virulence and pathogenicity, covering its anticipated structure, probable biological functions, and importance in this context.
Results
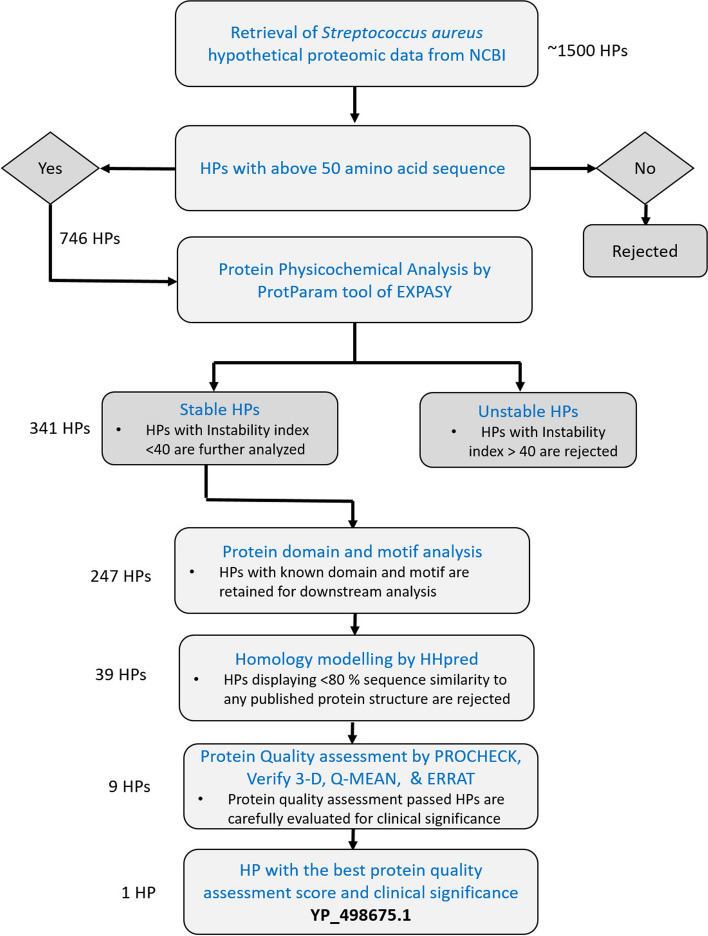
A hypothetical protein, YP_498675.1 with 281 amino acid residues of S. aureus, was chosen for analysis and modeling by several bioinformatics tools and databases in this work. According to primary and secondary structure analyses, YP_498675.1 is a stable hydrophilic protein with a significant proportion of α–helices. Subcellular localization predictions by CELLO, PSORTb, and SOSUI server indicate that it is a cytoplasmic protein. NCBI-CDD, Pfam, and InterProScan functional genomics research revealed that the hypothetical protein may include the pyridoxal phosphate (PLP)-dependent 2, 3-diaminopropionate biosynthesis protein SbnA domain. In the homology modeling method, the HHpred server was employed to create its 3D structure using the template structure of a Staphyloferrin B precursor biosynthetic enzyme SbnA bound to PLP (PDB ID: 5D84_A), an X-ray diffraction model having 100% sequence identity with the hypothetical protein. After energy minimization, several quality assessments and validation factors determined that the generated protein model was reliable and of reasonable quality.
Conclusion
The present study has characterized and functionally annotated the hypothetical protein YP_498675.1 of S. aureus. Further experimental validation would aid in determining the actual function of YP_498675.1 as well as confirm the protein’s value as a therapeutic target.


对来自金黄色葡萄球菌的一种新的假设蛋白(YP_498675.1)进行计算机分析,揭示了色氨酸合成酶β超家族的蛋白(Try-synth-beta_ II)。
背景:金黄色葡萄球菌是一种革兰氏阳性球形细菌,是世界上最常见的医院感染原因。鉴于其临床意义,金黄色葡萄球菌的基因组序列已被阐明,以提高我们对其生活方式和致病性的理解。本研究旨在总结一种可能在金黄色葡萄球菌毒力和致病性中发挥重要作用的潜在假设蛋白,包括其预期结构、可能的生物学功能及其在此背景下的重要性。结果:利用多种生物信息学工具和数据库,选择含有281个金黄色葡萄球菌氨基酸残基的假设蛋白YP_498675.1进行分析和建模。一级和二级结构分析表明,YP_498675.1是一种稳定的亲水性蛋白,α-螺旋比例显著。通过CELLO、PSORTb和SOSUI服务器的亚细胞定位预测表明它是一种细胞质蛋白。NCBI-CDD、Pfam和InterProScan功能基因组学研究显示,该假设蛋白可能包含磷酸吡哆醛(PLP)依赖性2,3 -二氨基丙酸生物合成蛋白SbnA结构域。在同源性建模方法中,利用与假设蛋白序列100%一致的x射线衍射模型,利用与PLP结合的Staphyloferrin B前体生物合成酶SbnA (PDB ID: 5D84_A)的模板结构,利用HHpred服务器创建其三维结构。在能量最小化之后,几个质量评估和验证因素确定了生成的蛋白质模型是可靠的,质量合理。结论:本研究对假设的金黄色葡萄球菌蛋白YP_498675.1进行了表征和功能注释。进一步的实验验证将有助于确定YP_498675.1的实际功能,并确认该蛋白作为治疗靶点的价值。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Genetic Engineering and Biotechnology
Biochemistry, Genetics and Molecular Biology-Biotechnology
CiteScore
5.70
自引率
5.70%
发文量
159
审稿时长
16 weeks
期刊介绍:
Journal of genetic engineering and biotechnology is devoted to rapid publication of full-length research papers that leads to significant contribution in advancing knowledge in genetic engineering and biotechnology and provide novel perspectives in this research area. JGEB includes all major themes related to genetic engineering and recombinant DNA. The area of interest of JGEB includes but not restricted to: •Plant genetics •Animal genetics •Bacterial enzymes •Agricultural Biotechnology, •Biochemistry, •Biophysics, •Bioinformatics, •Environmental Biotechnology, •Industrial Biotechnology, •Microbial biotechnology, •Medical Biotechnology, •Bioenergy, Biosafety, •Biosecurity, •Bioethics, •GMOS, •Genomic, •Proteomic JGEB accepts
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


