Analyzing CRISPR screens in non-conventional microbes.
IF 3.2
4区 生物学
Q2 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 1
Abstract
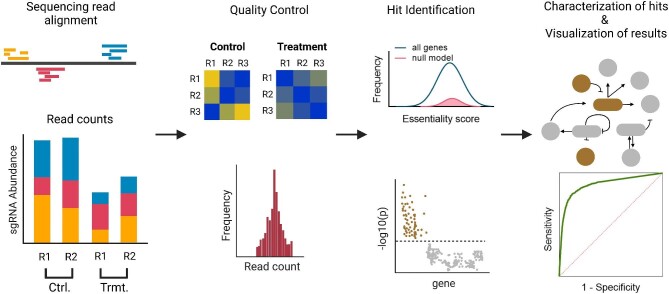
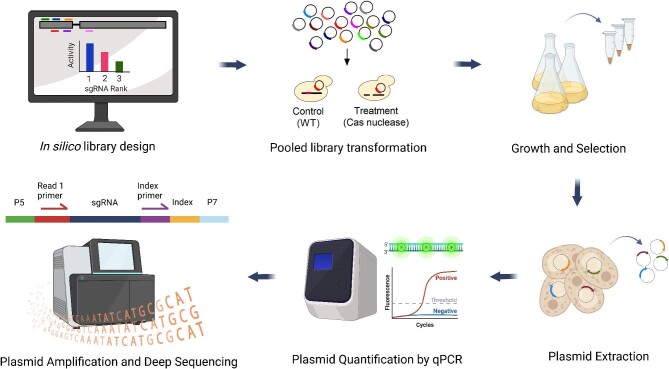
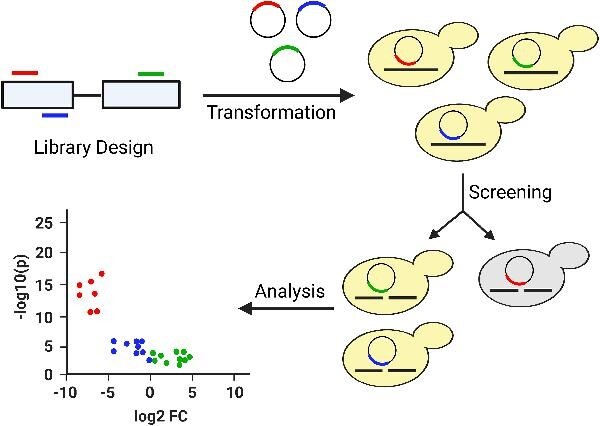
Abstract The multifaceted nature of CRISPR screens has propelled advancements in the field of functional genomics. Pooled CRISPR screens involve creating programmed genetic perturbations across multiple genomic sites in a pool of host cells subjected to a challenge, empowering researchers to identify genetic causes of desirable phenotypes. These genome-wide screens have been widely used in mammalian cells to discover biological mechanisms of diseases and drive the development of targeted drugs and therapeutics. Their use in non-model organisms, especially in microbes to improve bioprocessing-relevant phenotypes, has been limited. Further compounding this issue is the lack of bioinformatic algorithms for analyzing microbial screening data with high accuracy. Here, we describe the general approach and underlying principles for conducting pooled CRISPR knockout screens in non-conventional yeasts and performing downstream analysis of the screening data, while also reviewing state-of-the-art algorithms for identification of CRISPR screening outcomes. Application of pooled CRISPR screens to non-model yeasts holds considerable potential to uncover novel metabolic engineering targets and improve industrial bioproduction. One-Sentence Summary This mini-review describes experimental and computational approaches for functional genomic screening using CRISPR technologies in non-conventional microbes.
分析非常规微生物中的CRISPR筛选。
CRISPR筛选的多面性推动了功能基因组学领域的进步。汇集的CRISPR筛选涉及在宿主细胞池中受到挑战的多个基因组位点上创建程序化的遗传扰动,使研究人员能够确定理想表型的遗传原因。这些全基因组筛选已广泛应用于哺乳动物细胞中,以发现疾病的生物学机制,并推动靶向药物和治疗方法的发展。它们在非模式生物中的应用,特别是在微生物中改善生物加工相关表型的应用,一直受到限制。使这一问题进一步复杂化的是缺乏生物信息学算法来高精度地分析微生物筛选数据。在这里,我们描述了在非常规酵母中进行联合CRISPR敲除筛选和对筛选数据进行下游分析的一般方法和基本原则,同时也回顾了用于鉴定CRISPR筛选结果的最先进算法。将聚合CRISPR筛选应用于非模式酵母具有发现新的代谢工程靶点和改善工业生物生产的巨大潜力。一句话总结:这篇迷你综述描述了在非常规微生物中使用CRISPR技术进行功能基因组筛选的实验和计算方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Industrial Microbiology & Biotechnology
工程技术-生物工程与应用微生物
CiteScore
7.70
自引率
0.00%
发文量
25
审稿时长
3 months
期刊介绍:
The Journal of Industrial Microbiology and Biotechnology is an international journal which publishes papers describing original research, short communications, and critical reviews in the fields of biotechnology, fermentation and cell culture, biocatalysis, environmental microbiology, natural products discovery and biosynthesis, marine natural products, metabolic engineering, genomics, bioinformatics, food microbiology, and other areas of applied microbiology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


