Spatial epigenome–transcriptome co-profiling of mammalian tissues
IF 50.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 38
Abstract
Emerging spatial technologies, including spatial transcriptomics and spatial epigenomics, are becoming powerful tools for profiling of cellular states in the tissue context1–5. However, current methods capture only one layer of omics information at a time, precluding the possibility of examining the mechanistic relationship across the central dogma of molecular biology. Here, we present two technologies for spatially resolved, genome-wide, joint profiling of the epigenome and transcriptome by cosequencing chromatin accessibility and gene expression, or histone modifications (H3K27me3, H3K27ac or H3K4me3) and gene expression on the same tissue section at near-single-cell resolution. These were applied to embryonic and juvenile mouse brain, as well as adult human brain, to map how epigenetic mechanisms control transcriptional phenotype and cell dynamics in tissue. Although highly concordant tissue features were identified by either spatial epigenome or spatial transcriptome we also observed distinct patterns, suggesting their differential roles in defining cell states. Linking epigenome to transcriptome pixel by pixel allows the uncovering of new insights in spatial epigenetic priming, differentiation and gene regulation within the tissue architecture. These technologies are of great interest in life science and biomedical research. The authors present two technologies for spatially resolved, genome-wide, joint profiling of the epigenome and transcriptome by cosequencing chromatin accessibility and gene expression, or histone modifications and gene expression on the same tissue section at near-single-cell resolution.
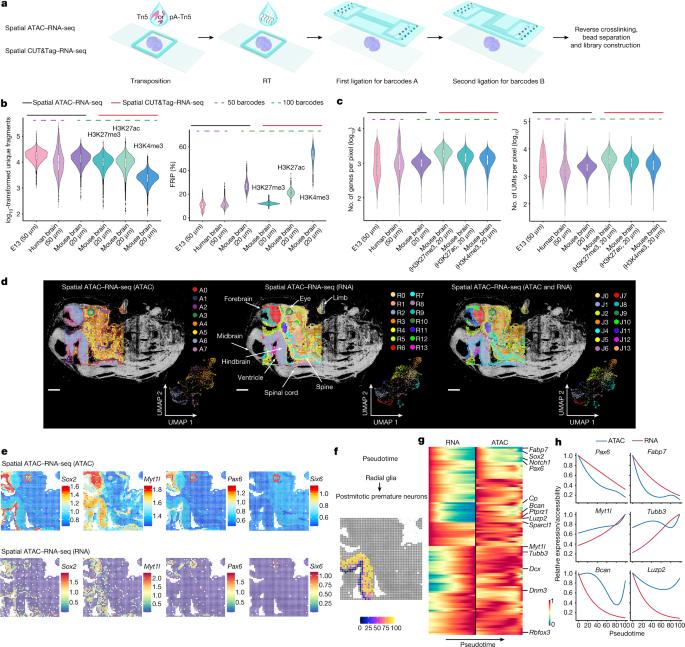
哺乳动物组织的空间表观基因组-转录组共谱分析。
新兴的空间技术,包括空间转录组学和空间表观基因组学,正在成为分析组织环境中细胞状态的有力工具1-5。然而,目前的方法一次只能捕获一层组学信息,排除了检查跨分子生物学中心教条的机制关系的可能性。在这里,我们提出了两种技术,通过染色质可及性和基因表达,或组蛋白修饰(H3K27me3, H3K27ac或H3K4me3)和基因表达在接近单细胞分辨率的同一组织切片上进行空间分辨,全基因组,表观基因组和转录组的联合分析。这些应用于胚胎和幼年小鼠大脑以及成人大脑,以绘制表观遗传机制如何控制组织中的转录表型和细胞动力学。虽然通过空间表观基因组或空间转录组鉴定出高度一致的组织特征,但我们也观察到不同的模式,表明它们在定义细胞状态方面的不同作用。将表观基因组与转录组逐像素连接,可以揭示组织结构中空间表观遗传启动、分化和基因调控的新见解。这些技术在生命科学和生物医学研究中具有重要意义。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


