A Combined Genomics and Phenomics Approach is Needed to Boost Breeding in Sugarcane.
IF 6.4
1区 农林科学
Q1 AGRONOMY
引用次数: 1
Abstract
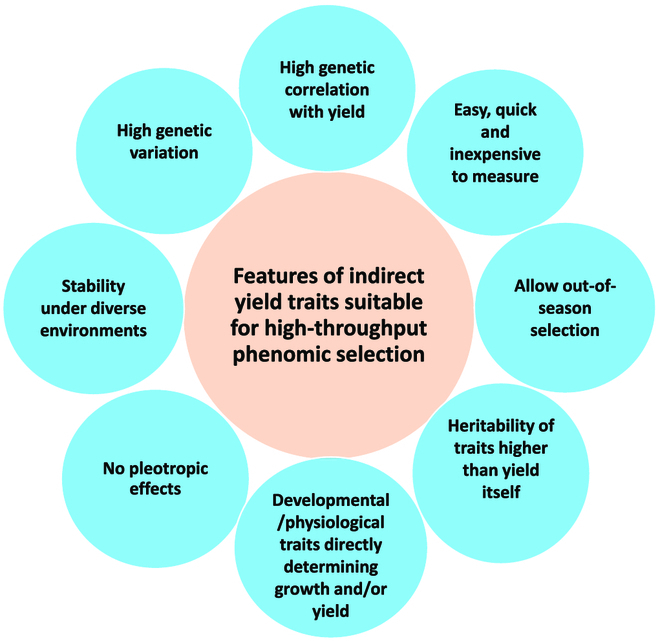
Sugarcane is a major food and bioenergy crop globally. It produces ~80% of sugar consumed worldwide, with Brazil and India together accounting for 61% of world sugarcane production in 2021 [1]. Globally, sugarcane is the 5th largest crop by production value and acreage, and it is also the second largest bioenergy crop [1,2]. Modern sugarcane is an interspecific hybrid (Saccharum species hybrid) of wild progenitor species Saccharum officinarum (2n = 80; x = 10) and Saccharum spontaneum (2n = 40 to 130; x = 8) [3]. This genetically complex polyploid crop with varied chromosome numbers (100 to 130) has one of the largest genomes (~10 kb) among plants, making sugarcane breeding considerably slow and challenging. Sugarcane breeding involves visual clonal selection combined with manual screening for cane stalk weight and cane sugar content through a 10to 12-year-long multistage selection scheme with disease screening incorporated toward the end of the selection program. Globally, the rate of sugarcane yield improvement realized at commercial crop production level through breeding in recent decades remains considerably lower than that of other major crops, and in some breeding programs, genetic gain appears to have plateaued [1]. Long breeding cycle, practical difficulties for extensive phenotyping of breeding populations, low narrow-sense heritability of economically important traits, large complex polyploid genome with high heterozygosity, and genotype–environment– management interaction effects have been attributed to low rate of genetic gain. More specifically, the high biomass of sugarcane plants makes accurate detailed phenotyping logistically very challenging, which compromises selection accuracy. This is particularly so in the early stages of selection confounded by large interplot competition effects caused by small singleor 2-row plots [4]. Thus, accurate, cost-effective, and high-throughput phenotyping offers an excellent opportunity for more precise estimation of true yield potential of sugarcane clones in breeding trials, a major bottleneck for fast-tracking sugarcane improvement [5]. Recognizing the persisting slow yield improvement from sugarcane breeding and the accelerated genetic gains realized through molecular marker-assisted selection (MAS) in various other crops [6,7], some of the leading sugarcane industries invested substantial resources for sugarcane genome sequencing and MAS in the past 3 decades [8]. Over this period, sugarcane DNA marker systems have gradually evolved from the initial hybridizationbased [9] to the current DNA-sequence-derived singlenucleotide polymorphism (SNP) markers, facilitated by high-throughput nextgeneration sequencing technologies [8]. The rapid advancements in DNA sequencing and marker technologies led to the creation of genotyping systems for wholegenome profiling, such as genomic selection (GS), which further strengthened marker discovery and marker-trait association studies. GS is a robust genotyping method capable of using large number of trait-linked DNA makers (e.g., SNP markers) spread across the whole genome and provides a more robust estimation of the genetic merit of a clone (for economically important traits) than previously achieved. Multiple independent studies found that it can be used as an effective high-throughput genetic screen for selecting elite sugarcane clones in breeding programs [8]. Using SNP markers, Deomano et al. [10], Yadav et al. [11], and, more recently, O’Connell et al. [12] proved the relative advantages of GS over conventional selection for cane yield, sugar content, and disease resistance. However, despite the long history of molecular marker discovery, application of MAS, including GS, in sugarcane breeding is yet to be realized. Reliable, accurate phenotyping of large genetically diverse genotyped populations on a regular basis is required to implement GS in sugarcane breeding. While experimental data and modeling have shown the predictive power (estimation of clone genetic merit) and potential variety development application of GS in sugarcane, its success in breeding for complex quantitative traits such as cane yield and sugar content, which are predominantly controlled by nonadditive genetic effects (i.e., interactions among genes or alleles that are not readily passed on from parents to progeny) [11], depends on, at least in part, accurate, high-throughput, and cost-effective phenotyping. High-throughput phenomics thus will greatly reduce the cost and increase the efficiency of creating reliable GS training sets Citation: Luo T, Liu X, Lakshmanan P. A Combined Genomics and Phenomics Approach is Needed to Boost Breeding in Sugarcane. Plant Phenomics 2023;5:Article 0074. https://doi.org/10.34133/ plantphenomics.0074
需要基因组学和表型组学相结合的方法来促进甘蔗育种。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Plant Phenomics
Multiple-
CiteScore
8.60
自引率
9.20%
发文量
26
审稿时长
14 weeks
期刊介绍:
Plant Phenomics is an Open Access journal published in affiliation with the State Key Laboratory of Crop Genetics & Germplasm Enhancement, Nanjing Agricultural University (NAU) and published by the American Association for the Advancement of Science (AAAS). Like all partners participating in the Science Partner Journal program, Plant Phenomics is editorially independent from the Science family of journals.
The mission of Plant Phenomics is to publish novel research that will advance all aspects of plant phenotyping from the cell to the plant population levels using innovative combinations of sensor systems and data analytics. Plant Phenomics aims also to connect phenomics to other science domains, such as genomics, genetics, physiology, molecular biology, bioinformatics, statistics, mathematics, and computer sciences. Plant Phenomics should thus contribute to advance plant sciences and agriculture/forestry/horticulture by addressing key scientific challenges in the area of plant phenomics.
The scope of the journal covers the latest technologies in plant phenotyping for data acquisition, data management, data interpretation, modeling, and their practical applications for crop cultivation, plant breeding, forestry, horticulture, ecology, and other plant-related domains.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


