Genome-wide analysis of a model-derived binge eating disorder phenotype identifies risk loci and implicates iron metabolism
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 1
Abstract
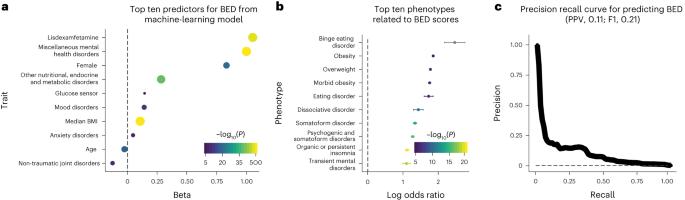
Binge eating disorder (BED) is the most common eating disorder, yet its genetic architecture remains largely unknown. Studying BED is challenging because it is often comorbid with obesity, a common and highly polygenic trait, and it is underdiagnosed in biobank data sets. To address this limitation, we apply a supervised machine-learning approach (using 822 cases of individuals diagnosed with BED) to estimate the probability of each individual having BED based on electronic medical records from the Million Veteran Program. We perform a genome-wide association study of individuals of African (n = 77,574) and European (n = 285,138) ancestry while controlling for body mass index to identify three independent loci near the HFE, MCHR2 and LRP11 genes and suggest APOE as a risk gene for BED. We identify shared heritability between BED and several neuropsychiatric traits, and implicate iron metabolism in the pathophysiology of BED. Overall, our findings provide insights into the genetics underlying BED and suggest directions for future translational research. Genome-wide association analysis of a binge eating disorder phenotype derived from a supervised machine-learning model applied to electronic medical records identifies three risk loci for this disorder and implicates iron metabolism in its etiology.
对模型衍生的暴饮症表型的全基因组分析确定了风险基因座,并暗示了铁代谢。
暴饮性进食障碍(BED)是最常见的进食障碍,但其遗传结构在很大程度上仍然未知。研究BED具有挑战性,因为它经常与肥胖合并,肥胖是一种常见的高度多基因特征,而且在生物库数据集中诊断不足。为了解决这一限制,我们应用了一种有监督的机器学习方法(使用822例被诊断为BED的个体),根据百万退伍军人计划的电子医疗记录来估计每个个体患BED的概率。我们对非洲(n = 77574)和欧洲(n = 285138)祖先,同时控制体重指数以确定HFE、MCHR2和LRP11基因附近的三个独立基因座并提示APOE是BED的风险基因。我们确定了BED和几种神经精神特征之间的共同遗传力,并将铁代谢与BED的病理生理学联系起来。总的来说,我们的发现为BED的遗传学基础提供了见解,并为未来的转化研究提供了方向。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


