Harnessing deep learning for population genetic inference
IF 39.1
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 1
Abstract
In population genetics, the emergence of large-scale genomic data for various species and populations has provided new opportunities to understand the evolutionary forces that drive genetic diversity using statistical inference. However, the era of population genomics presents new challenges in analysing the massive amounts of genomes and variants. Deep learning has demonstrated state-of-the-art performance for numerous applications involving large-scale data. Recently, deep learning approaches have gained popularity in population genetics; facilitated by the advent of massive genomic data sets, powerful computational hardware and complex deep learning architectures, they have been used to identify population structure, infer demographic history and investigate natural selection. Here, we introduce common deep learning architectures and provide comprehensive guidelines for implementing deep learning models for population genetic inference. We also discuss current challenges and future directions for applying deep learning in population genetics, focusing on efficiency, robustness and interpretability. Applying deep learning to large-scale genomic data of species or populations is providing new opportunities to understand the evolutionary forces that drive genetic diversity. This Review introduces common deep learning architectures and provides comprehensive guidelines to implement deep learning models for population genetic inference. The authors also discuss current opportunities and challenges for deep learning in population genetics.
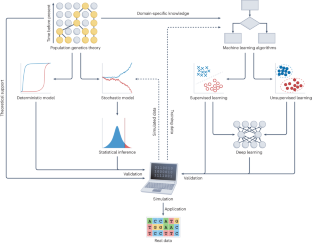
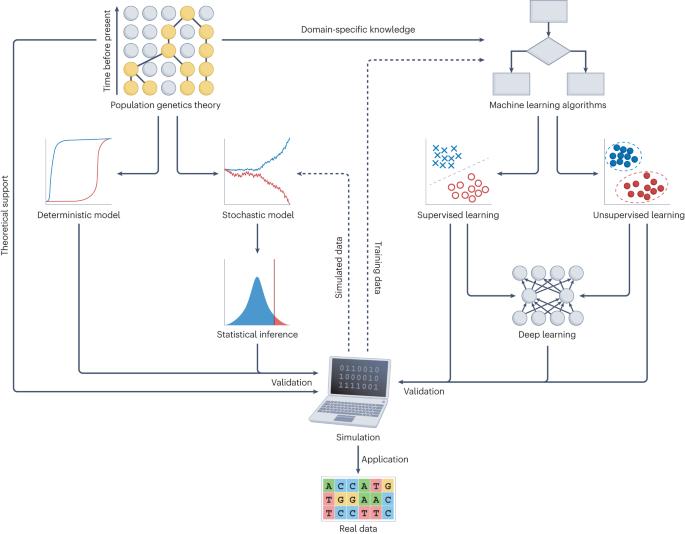
利用深度学习进行群体遗传推断。
在群体遗传学中,各种物种和种群的大规模基因组数据的出现为利用统计推断了解驱动遗传多样性的进化力量提供了新的机遇。然而,群体基因组学时代给分析海量基因组和变异带来了新的挑战。深度学习已在涉及大规模数据的众多应用中展示了最先进的性能。最近,深度学习方法在群体遗传学中大受欢迎;在海量基因组数据集、强大的计算硬件和复杂的深度学习架构的推动下,它们已被用于识别群体结构、推断人口历史和研究自然选择。在此,我们将介绍常见的深度学习架构,并为实施用于群体遗传推断的深度学习模型提供全面指导。我们还讨论了将深度学习应用于群体遗传学的当前挑战和未来方向,重点关注效率、鲁棒性和可解释性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Reviews Genetics
生物-遗传学
CiteScore
57.40
自引率
0.50%
发文量
113
审稿时长
6-12 weeks
期刊介绍:
At Nature Reviews Genetics, our goal is to be the leading source of reviews and commentaries for the scientific communities we serve. We are dedicated to publishing authoritative articles that are easily accessible to our readers. We believe in enhancing our articles with clear and understandable figures, tables, and other display items. Our aim is to provide an unparalleled service to authors, referees, and readers, and we are committed to maximizing the usefulness and impact of each article we publish.
Within our journal, we publish a range of content including Research Highlights, Comments, Reviews, and Perspectives that are relevant to geneticists and genomicists. With our broad scope, we ensure that the articles we publish reach the widest possible audience.
As part of the Nature Reviews portfolio of journals, we strive to uphold the high standards and reputation associated with this esteemed collection of publications.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


