Constructing synthetic biology workflows in the cloud
Abstract
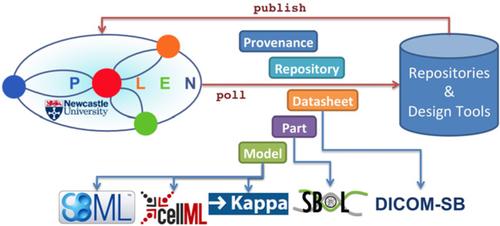
The synthetic biology design process has traditionally been heavily dependent upon manual searching, acquisition and integration of existing biological data. A large amount of such data is already available from Internet-based resources, but data exchange between these resources is often undertaken manually. Automating the communication between different resources can be done by the generation of computational workflows to achieve complex tasks that cannot be carried out easily or efficiently by a single resource. Computational workflows involve the passage of data from one resource, or process, to another in a distributed computing environment. In a typical bioinformatics workflow, the predefined order in which processes are invoked in a synchronous fashion and are described in a workflow definition document. However, in synthetic biology the diversity of resources and manufacturing tasks required favour a more flexible model for process execution. Here, the authors present the Protocol for Linking External Nodes (POLEN), a Cloud-based system that facilitates synthetic biology design workflows that operate asynchronously. Messages are used to notify POLEN resources of events in real time, and to log historical events such as the availability of new data, enabling networks of cooperation. POLEN can be used to coordinate the integration of different synthetic biology resources, to ensure consistency of information across distributed repositories through added support for data standards, and ultimately to facilitate the synthetic biology life cycle for designing and implementing biological systems.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


