Squid express conserved ADAR orthologs that possess novel features.
IF 4.4
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 1
Abstract
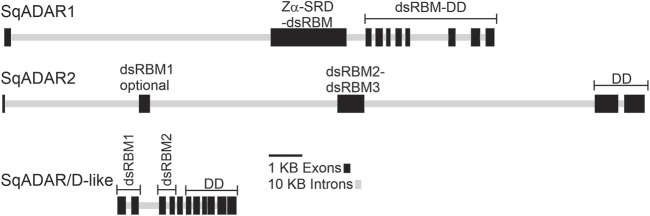
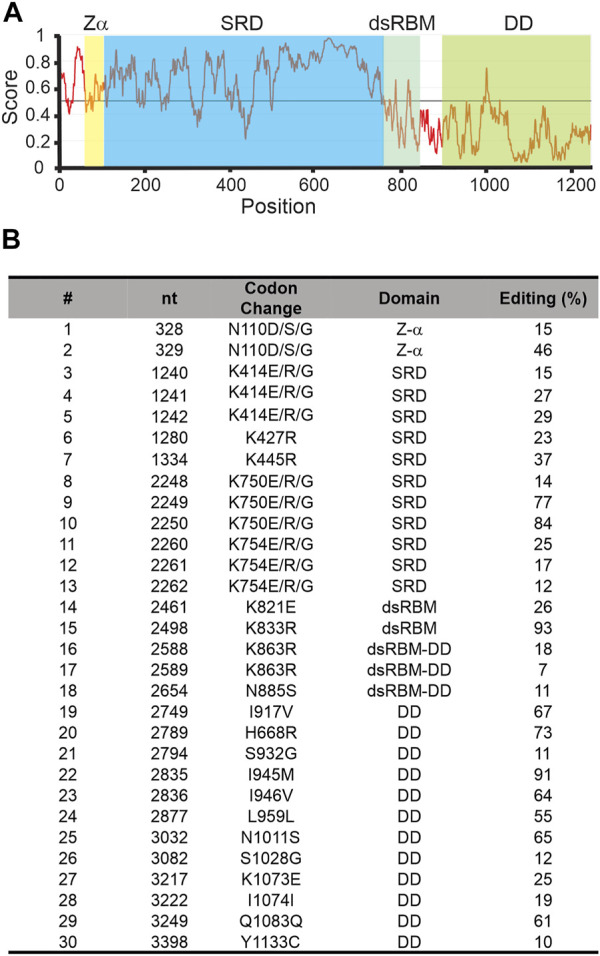
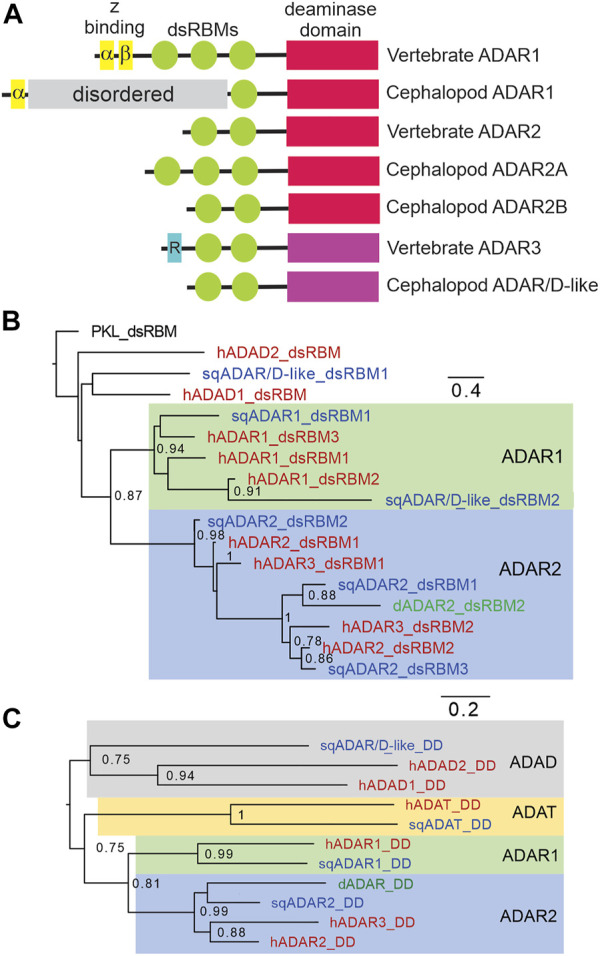
The coleoid cephalopods display unusually extensive mRNA recoding by adenosine deamination, yet the underlying mechanisms are not well understood. Because the adenosine deaminases that act on RNA (ADAR) enzymes catalyze this form of RNA editing, the structure and function of the cephalopod orthologs may provide clues. Recent genome sequencing projects have provided blueprints for the full complement of coleoid cephalopod ADARs. Previous results from our laboratory have shown that squid express an ADAR2 homolog, with two splice variants named sqADAR2a and sqADAR2b and that these messages are extensively edited. Based on octopus and squid genomes, transcriptomes, and cDNA cloning, we discovered that two additional ADAR homologs are expressed in coleoids. The first is orthologous to vertebrate ADAR1. Unlike other ADAR1s, however, it contains a novel N-terminal domain of 641 aa that is predicted to be disordered, contains 67 phosphorylation motifs, and has an amino acid composition that is unusually high in serines and basic amino acids. mRNAs encoding sqADAR1 are themselves extensively edited. A third ADAR-like enzyme, sqADAR/D-like, which is not orthologous to any of the vertebrate isoforms, is also present. Messages encoding sqADAR/D-like are not edited. Studies using recombinant sqADARs suggest that only sqADAR1 and sqADAR2 are active adenosine deaminases, both on perfect duplex dsRNA and on a squid potassium channel mRNA substrate known to be edited in vivo. sqADAR/D-like shows no activity on these substrates. Overall, these results reveal some unique features in sqADARs that may contribute to the high-level RNA recoding observed in cephalopods.
鱿鱼表达保守的ADAR同源物,具有新颖的特征。
胶质类头足类动物通过腺苷脱胺表现出异常广泛的mRNA重编码,但其潜在机制尚不清楚。由于作用于RNA (ADAR)酶的腺苷脱氨酶催化这种形式的RNA编辑,因此头足类同源动物的结构和功能可能提供线索。最近的基因组测序项目提供了完整的coleoid头足类ADARs的蓝图。我们实验室之前的结果表明,鱿鱼表达ADAR2同源物,具有两个剪接变体,称为sqADAR2a和sqADAR2b,这些信息被广泛编辑。基于章鱼和鱿鱼的基因组、转录组和cDNA克隆,我们发现了另外两个ADAR同源物在结肠中表达。第一个与脊椎动物ADAR1同源。然而,与其他adar1不同的是,它含有一个新的n端结构域641 aa,预计是无序的,包含67个磷酸化基序,并且具有异常高的丝氨酸和碱性氨基酸组成。编码sqADAR1的mrna本身被广泛编辑。第三种类似adar的酶,sqADAR/D-like,它与任何脊椎动物的同种异构体都不同源,也存在。编码为sqADAR/D-like的消息不被编辑。使用重组sqADARs的研究表明,只有sqADAR1和sqADAR2是活性腺苷脱氨酶,它们都位于完美双链dsRNA和已知在体内被编辑的鱿鱼钾通道mRNA底物上。sqADAR/D-like对这些底物无活性。总的来说,这些结果揭示了sqadar的一些独特特征,这些特征可能有助于在头足类动物中观察到的高水平RNA重新编码。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


